The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex
- PMID: 24748658
- PMCID: PMC4041447
- DOI: 10.1093/nar/gku274
The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex
Abstract
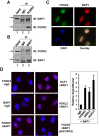
There are numerous forkhead transcription factors in mammalian cells but we know little about the molecular functions of the majority of these. FOXK2 is a ubiquitously expressed family member suggesting an important function across multiple cell types. Here, we show that FOXK2 binds to the SIN3A and PR-DUB complexes. The PR-DUB complex contains the important tumour suppressor protein, the deubiquitinase BAP1. FOXK2 recruits BAP1 to DNA, promotes local histone deubiquitination and causes changes in target gene activity. Our results therefore provide an important link between BAP1 and the transcription factor FOXK2 and demonstrate how BAP1 can be recruited to specific regulatory loci.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Tumor-derived neomorphic mutations in ASXL1 impairs the BAP1-ASXL1-FOXK1/K2 transcription network.Protein Cell. 2021 Jul;12(7):557-577. doi: 10.1007/s13238-020-00754-2. Epub 2020 Jul 18. Protein Cell. 2021. PMID: 32683582 Free PMC article.
-
BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes.J Biol Chem. 2015 Jan 16;290(3):1580-91. doi: 10.1074/jbc.M114.609834. Epub 2014 Dec 1. J Biol Chem. 2015. PMID: 25451922 Free PMC article.
-
PR-DUB maintains the expression of critical genes through FOXK1/2- and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination.Genome Res. 2020 Aug;30(8):1119-1130. doi: 10.1101/gr.261016.120. Epub 2020 Aug 3. Genome Res. 2020. PMID: 32747411 Free PMC article.
-
Roles and mechanisms of BAP1 deubiquitinase in tumor suppression.Cell Death Differ. 2021 Feb;28(2):606-625. doi: 10.1038/s41418-020-00709-4. Epub 2021 Jan 18. Cell Death Differ. 2021. PMID: 33462414 Free PMC article. Review.
-
An emerging model for BAP1's role in regulating cell cycle progression.Cell Biochem Biophys. 2011 Jun;60(1-2):3-11. doi: 10.1007/s12013-011-9184-6. Cell Biochem Biophys. 2011. PMID: 21484256 Free PMC article. Review.
Cited by
-
BAP1 shapes the bone marrow niche for lymphopoiesis by fine-tuning epigenetic profiles in endosteal mesenchymal stromal cells.Cell Death Differ. 2022 Nov;29(11):2151-2162. doi: 10.1038/s41418-022-01006-y. Epub 2022 Apr 26. Cell Death Differ. 2022. PMID: 35473985 Free PMC article.
-
BAP1-related signature predicts benefits from immunotherapy over VEGFR/mTOR inhibitors in ccRCC: a retrospective analysis of JAVELIN Renal 101 and checkmate-009/010/025 trials.Cancer Immunol Immunother. 2023 Aug;72(8):2557-2572. doi: 10.1007/s00262-023-03424-4. Epub 2023 Apr 12. Cancer Immunol Immunother. 2023. PMID: 37046008 Free PMC article.
-
Molecular architecture of polycomb repressive complexes.Biochem Soc Trans. 2017 Feb 8;45(1):193-205. doi: 10.1042/BST20160173. Biochem Soc Trans. 2017. PMID: 28202673 Free PMC article. Review.
-
FOXK2 transcription factor and its roles in tumorigenesis (Review).Oncol Lett. 2022 Nov 3;24(6):461. doi: 10.3892/ol.2022.13581. eCollection 2022 Dec. Oncol Lett. 2022. PMID: 36380871 Free PMC article. Review.
-
Tumor-derived neomorphic mutations in ASXL1 impairs the BAP1-ASXL1-FOXK1/K2 transcription network.Protein Cell. 2021 Jul;12(7):557-577. doi: 10.1007/s13238-020-00754-2. Epub 2020 Jul 18. Protein Cell. 2021. PMID: 32683582 Free PMC article.
References
-
- Carlsson P., Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 2002;250:1–23. - PubMed
-
- Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

