Histone H3 K79 methylation states play distinct roles in UV-induced sister chromatid exchange and cell cycle checkpoint arrest in Saccharomyces cerevisiae
- PMID: 24748660
- PMCID: PMC4041417
- DOI: 10.1093/nar/gku242
Histone H3 K79 methylation states play distinct roles in UV-induced sister chromatid exchange and cell cycle checkpoint arrest in Saccharomyces cerevisiae
Abstract
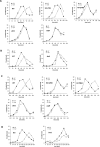
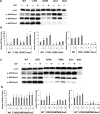
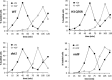
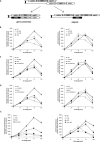
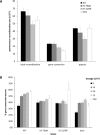
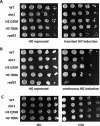
Histone post-translational modifications have been shown to contribute to DNA damage repair. Prior studies have suggested that specific H3K79 methylation states play distinct roles in the response to UV-induced DNA damage. To evaluate these observations, we examined the effect of altered H3K79 methylation patterns on UV-induced G1/S checkpoint response and sister chromatid exchange (SCE). We found that the di- and trimethylated states both contribute to activation of the G1/S checkpoint to varying degrees, depending on the synchronization method, although methylation is not required for checkpoint in response to high levels of UV damage. In contrast, UV-induced SCE is largely a product of the trimethylated state, which influences the usage of gene conversion versus popout mechanisms. Regulation of H3K79 methylation by H2BK123 ubiquitylation is important for both checkpoint function and SCE. H3K79 methylation is not required for the repair of double-stranded breaks caused by transient HO endonuclease expression, but does play a modest role in survival from continuous exposure. The overall results provide evidence for the participation of H3K79 methylation in UV-induced recombination repair and checkpoint activation, and further indicate that the di- and trimethylation states play distinct roles in these DNA damage response pathways.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Kupiec M. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat. Res. 2000;451:91–105. - PubMed
-
- Thoma F. Repair of UV lesions in nucleosomes—intrinsic properties and remodeling. DNA Repair (Amst) 2005;4:855–869. - PubMed
-
- Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, DC: ASM Press; 2006.
-
- Prakash S., Prakash L. Nucleotide excision repair in yeast. Mutat. Res. 2000;451:13–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

