The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs
- PMID: 24748666
- PMCID: PMC4041450
- DOI: 10.1093/nar/gku277
The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs
Abstract
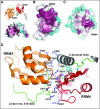
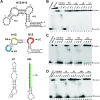
The Split Ends (SPEN) protein was originally discovered in Drosophila in the late 1990s. Since then, homologous proteins have been identified in eukaryotic species ranging from plants to humans. Every family member contains three predicted RNA recognition motifs (RRMs) in the N-terminal region of the protein. We have determined the crystal structure of the region of the human SPEN homolog that contains these RRMs-the SMRT/HDAC1 Associated Repressor Protein (SHARP), at 2.0 Å resolution. SHARP is a co-regulator of the nuclear receptors. We demonstrate that two of the three RRMs, namely RRM3 and RRM4, interact via a highly conserved interface. Furthermore, we show that the RRM3-RRM4 block is the main platform mediating the stable association with the H12-H13 substructure found in the steroid receptor RNA activator (SRA), a long, non-coding RNA previously shown to play a crucial role in nuclear receptor transcriptional regulation. We determine that SHARP association with SRA relies on both single- and double-stranded RNA sequences. The crystal structure of the SHARP-RRM fragment, together with the associated RNA-binding studies, extend the repertoire of nucleic acid binding properties of RRM domains suggesting a new hypothesis for a better understanding of SPEN protein functions.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

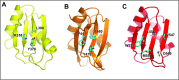

References
-
- Kolodziej P.A., Jan L.Y., Jan Y.N. Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron. 1995;15:273–286. - PubMed
-
- Wiellette E.L., Harding K.W., Mace K.A., Ronshaugen M.R., Wang F.Y., McGinnis W. Spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development. 1999;126:5373–5385. - PubMed
-
- Yabe D., Fukuda H., Aoki M., Yamada S., Takebayashi S., Shinkura R., Yamamoto N., Honjo T. Generation of a conditional knockout allele for mammalian Spen protein Mint/SHARP. Genesis. 2007;45:300–306. - PubMed
-
- Newberry E.P., Latifi T., Towler D.A. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry. 1999;38:10678–10690. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

