Do neonatal mouse hearts regenerate following heart apex resection?
- PMID: 24749066
- PMCID: PMC3986579
- DOI: 10.1016/j.stemcr.2014.02.008
Do neonatal mouse hearts regenerate following heart apex resection?
Abstract
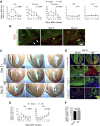
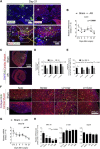
The mammalian heart has generally been considered nonregenerative, but recent progress suggests that neonatal mouse hearts have a genuine capacity to regenerate following apex resection (AR). However, in this study, we performed AR or sham surgery on 400 neonatal mice from inbred and outbred strains and found no evidence of complete regeneration. Ideally, new functional cardiomyocytes, endothelial cells, and vascular smooth muscle cells should be formed in the necrotic area of the damaged heart. Here, damaged hearts were 9.8% shorter and weighed 14% less than sham controls. In addition, the resection border contained a massive fibrotic scar mainly composed of nonmyocytes and collagen disposition. Furthermore, there was a substantial reduction in the number of proliferating cardiomyocytes in AR hearts. Our results thus question the usefulness of the AR model for identifying molecular mechanisms underlying regeneration of the adult heart after damage.
Figures


Comment in
-
Comment on "Do neonatal mouse hearts regenerate following heart apex resection"?Stem Cell Reports. 2014 Jul 8;3(1):2. doi: 10.1016/j.stemcr.2014.06.010. eCollection 2014 Jul 8. Stem Cell Reports. 2014. PMID: 25068115 Free PMC article. No abstract available.
-
Response to Sadek et al. and Kotlikoff et al.Stem Cell Reports. 2014 Jul 8;3(1):3-4. doi: 10.1016/j.stemcr.2014.06.011. eCollection 2014 Jul 8. Stem Cell Reports. 2014. PMID: 25068116 Free PMC article. No abstract available.
References
-
- Aguirre A., Sancho-Martinez I., Izpisua Belmonte J.C. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell. 2013;12:275–284. - PubMed
-
- Andersen D.C., Petersson S.J., Jørgensen L.H., Bollen P., Jensen P.B., Teisner B., Schroeder H.D., Jensen C.H. Characterization of DLK1+ cells emerging during skeletal muscle remodeling in response to myositis, myopathies, and acute injury. Stem Cells. 2009;27:898–908. - PubMed
-
- Drenckhahn J.D., Schwarz Q.P., Gray S., Laskowski A., Kiriazis H., Ming Z., Harvey R.P., Du X.J., Thorburn D.R., Cox T.C. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell. 2008;15:521–533. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

