Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor
- PMID: 24752081
- PMCID: PMC4356947
- DOI: 10.1038/nbt.2886
Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor
Abstract
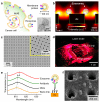
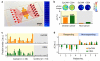
Exosomes show potential for cancer diagnostics because they transport molecular contents of the cells from which they originate. Detection and molecular profiling of exosomes is technically challenging and often requires extensive sample purification and labeling. Here we describe a label-free, high-throughput approach for quantitative analysis of exosomes. Our nano-plasmonic exosome (nPLEX) assay is based on transmission surface plasmon resonance through periodic nanohole arrays. Each array is functionalized with antibodies to enable profiling of exosome surface proteins and proteins present in exosome lysates. We show that this approach offers improved sensitivity over previous methods, enables portable operation when integrated with miniaturized optics and allows retrieval of exosomes for further study. Using nPLEX to analyze ascites samples from ovarian cancer patients, we find that exosomes derived from ovarian cancer cells can be identified by their expression of CD24 and EpCAM, suggesting the potential of exosomes for diagnostics.
Figures


Comment in
-
Tumor signatures in the blood.Nat Biotechnol. 2014 May;32(5):441-3. doi: 10.1038/nbt.2897. Nat Biotechnol. 2014. PMID: 24811515 No abstract available.
References
-
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. - PubMed
-
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. 2012;1820:940–948. - PubMed
-
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell. Biol. 2006 Chapter 3, Unit 3.22. - PubMed
-
- Brolo AG. Plasmonics for future biosensors. Nat. Photonics. 2012;6:709–713.
-
- Gordon R, Sinton D, Kavanagh KL, Brolo AG. A new generation of sensors based on extraordinary optical transmission. Acc. Chem. Res. 2008;41:1049–1057. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-HL113156/HL/NHLBI NIH HHS/United States
- P01 CA069246/CA/NCI NIH HHS/United States
- T32 CA079443/CA/NCI NIH HHS/United States
- HHSN268201000044C/HL/NHLBI NIH HHS/United States
- R01 EB010011/EB/NIBIB NIH HHS/United States
- R01-EB010011/EB/NIBIB NIH HHS/United States
- R01 CA204019/CA/NCI NIH HHS/United States
- T32CA79443/CA/NCI NIH HHS/United States
- K12 CA087723/CA/NCI NIH HHS/United States
- R01-EB00462605A1/EB/NIBIB NIH HHS/United States
- K12CA087723-11A1/CA/NCI NIH HHS/United States
- R01 EB004626/EB/NIBIB NIH HHS/United States
- R01 HL113156/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

