Mutated tumor alleles are expressed according to their DNA frequency
- PMID: 24752137
- PMCID: PMC3994436
- DOI: 10.1038/srep04743
Mutated tumor alleles are expressed according to their DNA frequency
Abstract
The transcription of tumor mutations from DNA into RNA has implications for biology, epigenetics and clinical practice. It is not clear if mutations are in general transcribed and, if so, at what proportion to the wild-type allele. Here, we examined the correlation between DNA mutation allele frequency and RNA mutation allele frequency. We sequenced the exome and transcriptome of tumor cell lines with large copy number variations, identified heterozygous single nucleotide mutations and absolute DNA copy number, and determined the corresponding DNA and RNA mutation allele fraction. We found that 99% of the DNA mutations in expressed genes are expressed as RNA. Moreover, we found a high correlation between the DNA and RNA mutation allele frequency. Exceptions are mutations that cause premature termination codons and therefore activate nonsense-mediated decay. Beyond this, we did not find evidence of any wide-scale mechanism, such as allele-specific epigenetic silencing, preferentially promoting mutated or wild-type alleles. In conclusion, our data strongly suggest that genes are equally transcribed from all alleles, mutated and wild-type, and thus transcribed in proportion to their DNA allele frequency.
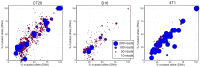
Figures



References
-
- Sjoblom T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006). - PubMed
-
- Allegra C. J. et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27, 2091–2096 (2009). - PubMed
-
- Chang Y. F., Imam J. S. & Wilkinson M. F. The nonsense-mediated decay RNA surveillance pathway. Annual review of biochemistry 76, 51–74 (2007). - PubMed
-
- Cartegni L., Chew S. L. & Krainer A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3, 285–298 (2002). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

