IncH-type plasmid harboring bla CTX-M-15, bla DHA-1, and qnrB4 genes recovered from animal isolates
- PMID: 24752252
- PMCID: PMC4068538
- DOI: 10.1128/AAC.02695-14
IncH-type plasmid harboring bla CTX-M-15, bla DHA-1, and qnrB4 genes recovered from animal isolates
Abstract
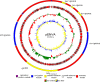
The whole sequence of plasmid pENVA carrying the extended-spectrum β-lactamase gene blaCTX-M-15 was determined. It was identified from a series of clonally related Klebsiella pneumoniae sequence type 274 strains recovered from companion animals. This plasmid was 253,984 bp in size and harbored, in addition to blaCTX-M-15, a large array of genes encoding resistance to many antibiotic molecules, including β-lactams (blaTEM-1, blaDHA-1), aminoglycosides (aacA2, aadA1), tetracycline (tetA), quinolones (qnrB4), trimethoprim (dfrA15), and sulfonamides (two copies of sul1). In addition, genes encoding resistance to mercury, tellurium, nickel, and quaternary compounds were identified. It also carried genes encoding DNA damage protection and mutagenesis repair and a locus for a CRISPR system, which corresponds to an immune system involved in protection against bacteriophages and plasmids. Comparative analysis of the plasmid scaffold showed that it possessed a structure similar to that of only a single plasmid, which was pNDM-MAR encoding the carbapenemase NDM-1 and identified from human K. pneumoniae isolates. Both plasmids possessed two replicons, namely, those of IncFIB-like and IncHIB-like plasmids, which were significantly different from those previously characterized. The blaCTX-M-15 gene, together with the other antibiotic resistance genes, was part of a large module likely acquired through a transposition process. We characterized here a new plasmid type carrying the blaCTX-M-15 gene identified in a K. pneumoniae isolate of animal origin. The extent to which this plasmid type may spread efficiently and possibly further enhance the dissemination of blaCTX-M-15 among animal and human isolates remains to be determined.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D, Peixe L, Poirel L, Schuepbach-Regula G, Torneke K, Torren-Edo J, Torres C, Threlfall J. 2013. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56:1030–1037. 10.1093/cid/cis1043 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

