Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress
- PMID: 24752898
- PMCID: PMC4054315
- DOI: 10.1128/MCB.00136-14
Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress
Abstract
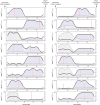
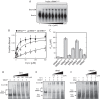
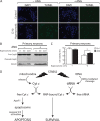
Adaptation to changes in extracellular tonicity is essential for cell survival. However, severe or chronic hyperosmotic stress induces apoptosis, which involves cytochrome c (Cyt c) release from mitochondria and subsequent apoptosome formation. Here, we show that angiogenin-induced accumulation of tRNA halves (or tiRNAs) is accompanied by increased survival in hyperosmotically stressed mouse embryonic fibroblasts. Treatment of cells with angiogenin inhibits stress-induced formation of the apoptosome and increases the interaction of small RNAs with released Cyt c in a ribonucleoprotein (Cyt c-RNP) complex. Next-generation sequencing of RNA isolated from the Cyt c-RNP complex reveals that 20 tiRNAs are highly enriched in the Cyt c-RNP complex. Preferred components of this complex are 5' and 3' tiRNAs of specific isodecoders within a family of isoacceptors. We also demonstrate that Cyt c binds tiRNAs in vitro, and the pool of Cyt c-interacting RNAs binds tighter than individual tiRNAs. Finally, we show that angiogenin treatment of primary cortical neurons exposed to hyperosmotic stress also decreases apoptosis. Our findings reveal a connection between angiogenin-generated tiRNAs and cell survival in response to hyperosmotic stress and suggest a novel cellular complex involving Cyt c and tiRNAs that inhibits apoptosome formation and activity.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
