GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state
- PMID: 24753578
- PMCID: PMC4020072
- DOI: 10.1073/pnas.1401706111
GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state
Abstract
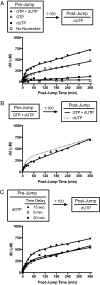
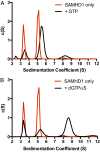
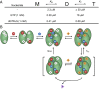
The HIV-1 restriction factor sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) is a tetrameric protein that catalyzes the hydrolysis of all dNTPs to the deoxynucleoside and tripolyphosphate, which effectively depletes the dNTP substrates of HIV reverse transcriptase. Here, we establish that SAMHD1 is activated by GTP binding to guanine-specific activator sites (A1) as well as coactivation by substrate dNTP binding to a distinct set of nonspecific activator sites (A2). Combined activation by GTP and dNTPs results in a long-lived tetrameric form of SAMHD1 that persists for hours, even after activating nucleotides are withdrawn from the solution. These results reveal an ordered model for assembly of SAMHD1 tetramer from its inactive monomer and dimer forms, where GTP binding to the A1 sites generates dimer and dNTP binding to the A2 and catalytic sites generates active tetramer. Thus, cellular regulation of active SAMHD1 is not determined by GTP alone but instead, the levels of all dNTPs and the generation of a persistent tetramer that is not in equilibrium with free activators. The significance of the long-lived activated state is that SAMHD1 can remain active long after dNTP pools have been reduced to a level that would lead to inactivation. This property would be important in resting CD4(+) T cells, where dNTP pools are reduced to nanomolar levels to restrict infection by HIV-1.
Keywords: dNTP induced oligomerization; enzyme catalysis; innate immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 against 1/[GTP]. (E) GTP concentration dependence of the dUTP hydrolysis rates at increasing [dUTP]. (F) Secondary plot of
against 1/[GTP]. (E) GTP concentration dependence of the dUTP hydrolysis rates at increasing [dUTP]. (F) Secondary plot of  against [dUTP].
against [dUTP].

References
-
- Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem. 1992;267(6):3589–3596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

