The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once
- PMID: 24754999
- PMCID: PMC4051888
- DOI: 10.1016/j.dnarep.2014.03.022
The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once
Abstract
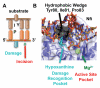
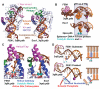
To avoid genome instability, DNA repair nucleases must precisely target the correct damaged substrate before they are licensed to incise. Damage identification is a challenge for all DNA damage response proteins, but especially for nucleases that cut the DNA and necessarily create a cleaved DNA repair intermediate, likely more toxic than the initial damage. How do these enzymes achieve exquisite specificity without specific sequence recognition or, in some cases, without a non-canonical DNA nucleotide? Combined structural, biochemical, and biological analyses of repair nucleases are revealing their molecular tools for damage verification and safeguarding against inadvertent incision. Surprisingly, these enzymes also often act on RNA, which deserves more attention. Here, we review protein-DNA structures for nucleases involved in replication, base excision repair, mismatch repair, double strand break repair (DSBR), and telomere maintenance: apurinic/apyrimidinic endonuclease 1 (APE1), Endonuclease IV (Nfo), tyrosyl DNA phosphodiesterase (TDP2), UV Damage endonuclease (UVDE), very short patch repair endonuclease (Vsr), Endonuclease V (Nfi), Flap endonuclease 1 (FEN1), exonuclease 1 (Exo1), RNase T and Meiotic recombination 11 (Mre11). DNA and RNA structure-sensing nucleases are essential to life with roles in DNA replication, repair, and transcription. Increasingly these enzymes are employed as advanced tools for synthetic biology and as targets for cancer prognosis and interventions. Currently their structural biology is most fully illuminated for DNA repair, which is also essential to life. How DNA repair enzymes maintain genome fidelity is one of the DNA double helix secrets missed by James Watson and Francis Crick, that is only now being illuminated though structural biology and mutational analyses. Structures reveal motifs for repair nucleases and mechanisms whereby these enzymes follow the old carpenter adage: measure twice, cut once. Furthermore, to measure twice these nucleases act as molecular level transformers that typically reshape the DNA and sometimes themselves to achieve extraordinary specificity and efficiency.
Keywords: APE1; Base excision repair; Crystallography; DNA; DNA repair; DNase; Double strand break repair; EndoIV; EndoV; Endonucleases; Enzyme–DNA complex; Exo1; Exonuclease; FEN1; Genome maintenance; Magnesium; Manganese; Metals; Mismatch repair; Mre11; Nfi; Nfo; Nucleases; Nucleotide incision repair; RNA; RNase; Structure-specific nuclease; TDP2; Telomere; UVDE; Vsr; Zinc.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures





References
-
- Crick F. The double helix: a personal view. Nature. 1974;248:766–769. - PubMed
-
- Arber W, Linn S. DNA modification and restriction. Annual Review of Biochemistry. 1969;38:467–500. - PubMed
-
- Harrington JJ, Lieber MR. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

