Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET
- PMID: 24755590
- PMCID: PMC4430801
- DOI: 10.1038/nchem.1919
Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET
Erratum in
- Nat Chem. 2014 Feb;7(2):178. Wilf, Nabil W [corrected to Wilf, Nabil M]
Abstract
The ability to introduce different biophysical probes into defined positions in target proteins will provide powerful approaches for interrogating protein structure, function and dynamics. However, methods for site-specifically incorporating multiple distinct unnatural amino acids are hampered by their low efficiency. Here we provide a general solution to this challenge by developing an optimized orthogonal translation system that uses amber and evolved quadruplet-decoding transfer RNAs to encode numerous pairs of distinct unnatural amino acids into a single protein expressed in Escherichia coli with a substantial increase in efficiency over previous methods. We also provide a general strategy for labelling pairs of encoded unnatural amino acids with different probes via rapid and spontaneous reactions under physiological conditions. We demonstrate the utility of our approach by genetically directing the labelling of several pairs of sites in calmodulin with fluorophores and probing protein structure and dynamics by Förster resonance energy transfer.
Figures



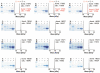
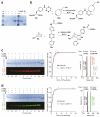
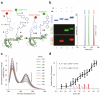
Comment in
-
Synthetic biology: Two-for-one designer labels.Nat Chem. 2014 May;6(5):379-81. doi: 10.1038/nchem.1935. Nat Chem. 2014. PMID: 24755585 No abstract available.
References
-
- Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nature Reviews Molecular Cell Biology. 2002;3:906–918. - PubMed
-
- Kajihara D, et al. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat Methods. 2006;3:923–929. - PubMed
-
- Li P, Roller PP. Cyclization strategies in peptide derived drug design. Curr Top Med Chem. 2002;2:325–341. - PubMed
-
- Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: from triplet to quadruplet codes. Angew Chem Int Ed Engl. 2012;51:2288–2297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

