Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization
- PMID: 24755653
- PMCID: PMC3995651
- DOI: 10.1371/journal.pone.0093060
Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization
Abstract
The regulation of membrane shapes is central to many cellular phenomena. Bin/Amphiphysin/Rvs (BAR) domain-containing proteins are key players for membrane remodeling during endocytosis, cell migration, and endosomal sorting. BIN1, which contains an N-BAR domain, is assumed to be essential for biogenesis of plasma membrane invaginations (T-tubules) in muscle tissues. Three mutations, K35N, D151N and R154Q, have been discovered so far in the BAR domain of BIN1 in patients with centronuclear myopathy (CNM), where impaired organization of T-tubules has been reported. However, molecular mechanisms behind this malfunction have remained elusive. None of the BIN1 disease mutants displayed a significantly compromised curvature sensing ability. However, two mutants showed impaired membrane tubulation both in vivo and in vitro, and displayed characteristically different behaviors. R154Q generated smaller membrane curvature compared to WT N-BAR. Quantification of protein density on membranes revealed a lower membrane-bound density for R154Q compared to WT and the other mutants, which appeared to be the primary reason for the observation of impaired deformation capacity. The D151N mutant was unable to tubulate liposomes under certain experimental conditions. At medium protein concentrations we found 'budding' structures on liposomes that we hypothesized to be intermediates during the tubulation process except for the D151N mutant. Chemical crosslinking assays suggested that the D151N mutation impaired protein oligomerization upon membrane binding. Although we found an insignificant difference between WT and K35N N-BAR in in vitro assays, depolymerizing actin in live cells allowed tubulation of plasma membranes through the K35N mutant. Our results provide insights into the membrane-involved pathophysiological mechanisms leading to human disease.
Conflict of interest statement
Figures





 /
/
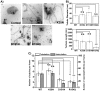
 from images recorded by Kalman-averaged confocal xz line-scan images is plotted with square root of membrane tension, revealing a linear relationship. Data from six vesicles was binned (vertical error bars represent standard error of the mean of
from images recorded by Kalman-averaged confocal xz line-scan images is plotted with square root of membrane tension, revealing a linear relationship. Data from six vesicles was binned (vertical error bars represent standard error of the mean of  /
/
 and horizontal error bars show standard error of the mean of square root of tension). Inset graph demonstrates similar fluorescence intensity on GUVs analyzed for four BIN1 variants. Error bars are standard errors of the mean. D) Equilibrium force of tethers (pulled from GUVs with composition (60% DOPC, 20% DOPS, 10% DOPE, 10% PI(4,5)P2) measured by optical trap at membrane tension of 0.21±0.003 mN/m. Error bars: standard error of the mean in black and standard deviation in light grey. Student t-test for statistical significance: n.s: p>0.05, *: p<0.05, **: p<0.01, ***: p<0.001.
and horizontal error bars show standard error of the mean of square root of tension). Inset graph demonstrates similar fluorescence intensity on GUVs analyzed for four BIN1 variants. Error bars are standard errors of the mean. D) Equilibrium force of tethers (pulled from GUVs with composition (60% DOPC, 20% DOPS, 10% DOPE, 10% PI(4,5)P2) measured by optical trap at membrane tension of 0.21±0.003 mN/m. Error bars: standard error of the mean in black and standard deviation in light grey. Student t-test for statistical significance: n.s: p>0.05, *: p<0.05, **: p<0.01, ***: p<0.001.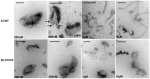




References
-
- McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438: 590–596. - PubMed
-
- Bhatia VK, Hatzakis NS, Stamou D (2010) A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Seminars in Cell & Developmental Biology 21: 381–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

