Derivation and characterization of Dicer- and microRNA-deficient human cells
- PMID: 24757167
- PMCID: PMC4024645
- DOI: 10.1261/rna.044545.114
Derivation and characterization of Dicer- and microRNA-deficient human cells
Abstract
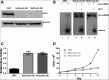
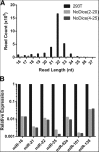
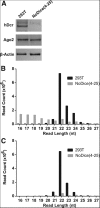
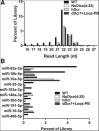
We have used genome editing to generate inactivating deletion mutations in all three copies of the dicer (hdcr) gene present in the human cell line 293T. As previously shown in murine ES cells lacking Dicer function, hDcr-deficient 293T cells are severely impaired for the production of mature microRNAs (miRNAs). Nevertheless, RNA-induced silencing complexes (RISCs) present in these hDcr-deficient cells are readily programmed by transfected, synthetic miRNA duplexes to repress mRNAs bearing either fully or partially complementary targets, including targets bearing incomplete seed homology to the introduced miRNA. Using these hDcr-deficient 293T cells, we demonstrate that human pre-miRNA processing can be effectively rescued by ectopic expression of the Drosophila Dicer 1 protein, but only in the presence of the PB isoform of Loquacious (Loqs-PB), the fly homolog of the hDcr cofactor TRBP. In contrast, Drosophila Dicer 2, even in the presence of its cofactors Loqs-PD and R2D2, was unable to support human pre-miRNA processing. Interestingly, although ectopic Drosophila Dicer 1/Loqs-PB or hDcr both rescued pre-miRNA processing effectively in these hDcr-deficient cells, there were significant differences in the ratio of the miRNA isoforms that were produced, especially in the case of miR-30 family members, and we also noted differences in the relative expression level of miRNAs vs. passenger strands for a subset of human miRNAs. These data demonstrate that the mechanisms underlying the accurate processing of pre-miRNAs are largely, but not entirely, conserved between mammalian and insect cells.
Keywords: Dicer; RISC; RNA interference; microRNAs; post-transcriptional regulation.
© 2014 Bogerd et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
