Amplification of aminoglycoside resistance gene aphA1 in Acinetobacter baumannii results in tobramycin therapy failure
- PMID: 24757213
- PMCID: PMC3994513
- DOI: 10.1128/mBio.00915-14
Amplification of aminoglycoside resistance gene aphA1 in Acinetobacter baumannii results in tobramycin therapy failure
Abstract
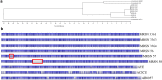
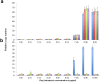
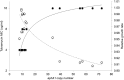
Gene amplification is believed to play an important role in antibiotic resistance but has been rarely documented in clinical settings because of its unstable nature. We report a rise in MICs from 0.5 to 16 μg/ml in successive Acinetobacter baumannii isolated over 4 days from a patient being treated with tobramycin for an infection by multidrug-resistant A. baumannii, resulting in therapeutic failure. Isolates were characterized by whole-genome sequencing, real-time and reverse transcriptase PCR, and growth assays to determine the mechanism of tobramycin resistance and its fitness cost. Tobramycin resistance was associated with two amplification events of different chromosomal fragments containing the aphA1 aminoglycoside resistance gene part of transposon Tn6020. The first amplification event involved low amplification (6 to 10 copies) of a large DNA fragment that was unstable and conferred tobramycin MICs of ≤ 8 μg/ml. The second event involved moderate (10 to 30 copies) or high (40 to 110 copies) amplification of Tn6020. High copy numbers were associated with tobramycin MICs of 16 μg/ml, impaired fitness, and genetic instability, whereas lower copy numbers resulted in tobramycin MICs of ≤8 μg/ml and no fitness cost and were stably maintained in vitro. Exposure in vitro to tobramycin of the initial susceptible isolate and of the A. baumannii AB0057 reference strain led to similar aphA1 amplifications and elevated tobramycin MICs. To the best of our knowledge, this is the first report of in vivo development of antibiotic resistance secondary to gene amplifications resulting in therapy failure. IMPORTANCE A combination of whole-genome sequencing and mapping were used to detect an antibiotic resistance mechanism, gene amplification, which has been presumed for a long time to be of major importance but has rarely been reported in clinical settings because of its unstable nature. Two gene amplification events in a patient with an Acinetobacter baumannii infection treated with tobramycin were identified. One gene amplification event led to high levels of resistance and was rapidly reversible, while the second event led to low and more stable resistance since it incurred low fitness cost on the host. Gene amplification, with an associated rise in tobramycin MICs, could be readily reproduced in vitro from initially susceptible strains exposed to increasing concentrations of tobramycin, suggesting that gene amplification in A. baumannii may be a more common mechanism than currently believed. This report underscores the importance of rapid molecular techniques for surveillance of drug resistance.
Figures


References
-
- Lesho E, Yoon EJ, McGann P, Snesrud E, Kwak Y, Milillo M, Onmus-Leone F, Preston L, St. Clair K, Nikolich M, Viscount H, Wortmann G, Zapor M, Grillot-Courvalin C, Courvalin P, Clifford R, Waterman PE. 2013. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J. Infect. Dis. 208:1142–1151. 10.1093/infdis/jit293 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
