HCV core antigen and HCV-RNA in HIV/HCV co-infected patients with different HCV genotypes
- PMID: 24758157
- PMCID: PMC4029812
- DOI: 10.1186/1471-2334-14-222
HCV core antigen and HCV-RNA in HIV/HCV co-infected patients with different HCV genotypes
Abstract
Background: A good correlation between HCV core antigen (HCVAg) and different HCV-RNA assays has been described, but little data are available in HCV/HIV co-infection. We aimed to evaluate HCVAg in comparison with HCV-RNA and to determine their kinetics during antiviral treatment in selected HCV/HIV co-infected patients.
Methods: 355 samples from 286 HCV/HIV co-infected subjects for whom HCV-RNA (Abbott RealTime) was requested were analysed also for HCVAg (Abbott ARCHITECT) in order to evaluate the correlation between the two parameters both in patients treated or untreated for chronic hepatitis C and according to different HCV genotypes. The differences between percentages were evaluated by chi square or Fisher's exact test, while mean and median values were compared by Student's t test or the Mann-Whitney test, respectively. All differences were considered significant for a p value <0.05.
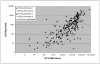
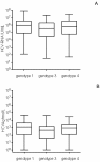
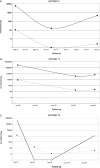
Results: HCVAg was detectable on 288/315 sera (91.4%) positive for HCV-RNA and in 5 out of40 (12.5%) sera with undetectable HCV-RNA for a total concordance of 90.1%. The correlation was fair both in untreated (r = 0.742) and in treated (r = 0.881) patients and stronger for genotypes 1 and 4 than for genotype 3. Both HCV-RNA and HCVAg levels were significantly higher (p = 0.028 and p = 0.0098, respectively) in patients infected by genotype 1 than by genotype 3. The mean ratio of Log values between HCV-RNA (IU/mL) and HCVAg (fmol/liter) was 2.27 ± 1.09 in untreated and 2.20 ± 0.82 in treated patients (p = n.s.),consistent with a sensitivity of HCVAg corresponding to about 1,000 IU/mL of HCV-RNA, and ranged from 2.21 to 2.32 among HCV genotypes with no significant differences; five samples (1.4%; 2 genotype 1a or 1c, 3 genotype 3a) showed highly divergent values. The analysis of 18 monitoring profiles from patients treated with PEG-IFN and Ribavirin showed similar trends, except in one case in which relapse could be predicted by HCVAg and not by HCV-RNA.
Conclusion: These results suggest that HCVAg represents an adequate tool for determining an ongoing HCV infection also in HIV co-infected patients, with lower costs and faster turnaround time than HCV-RNA.
Figures
References
-
- Hatzakis A, Wait S, Bruix J, Buti M, Carballo M, Cavaleri M, Colombo M, Delarocque-Astagneau E, Dusheiko G, Esmat G, Esteban R, Goldberg D, Gore C, Lok ASF, Manns M, Marcellin P, Papatheodoridis G, Peterle A, Prati D, Piorkowsky N, Rizzetto M, Roudot-Thoraval F, Soriano V, Thomas HC, Thursz M, Valla D, van Damme P, Veldhuijzen IK, Wedemeyer H, Wiessing L, Zanetti AR, Janssen HLA. et al.The state of hepatitis B and C in Europe: report from the hepatitis B and C summit conference. J Viral Hepat. 2011;14(suppl. 1):1–16. - PubMed
-
- European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatitis C virus infection. J Hepatol. 2011;14:245–264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

