Severe hypoxia exerts parallel and cell-specific regulation of gene expression and alternative splicing in human mesenchymal stem cells
- PMID: 24758227
- PMCID: PMC4234502
- DOI: 10.1186/1471-2164-15-303
Severe hypoxia exerts parallel and cell-specific regulation of gene expression and alternative splicing in human mesenchymal stem cells
Abstract
Background: The endosteum of the bone marrow provides a specialized hypoxic niche that may serve to preserve the integrity, pluripotency, longevity and stemness of resident mesenchymal stem cells (MSCs). To explore the molecular genetic consequences of such a niche we subjected human (h) MSCs to a pO2 of 4 mmHg and analyzed global gene expression and alternative splicing (AS) by genome-exon microarray and RT-qPCR, and phenotype by western blot and immunostaining.
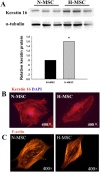
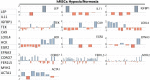
Results: Out of 446 genes differentially regulated by >2.5-fold, down-regulated genes outnumbered up-regulated genes by 243:203. Exon analyses revealed 60 hypoxia-regulated AS events with splice indices (SI) >1.0 from 53 genes and a correlation between high SI and degree of transcript regulation. Parallel analyses of a publicly available AS study on human umbilical vein endothelial cells (HUVECs) showed that there was a strong cell-specific component with only 11 genes commonly regulated in hMSCs and HUVECs and 17 common differentially spliced genes. Only 3 genes were differentially responsive to hypoxia at the gene (>2.0) and AS levels in both cell types. Functional assignments revealed unique profiles of gene expression with complex regulation of differentiation, extracellular matrix, intermediate filament and metabolic marker genes. Antioxidant genes, striated muscle genes and insulin/IGF-1 signaling intermediates were down-regulated. There was a coordinate induction of 9 out of 12 acidic keratins that along with other epithelial and cell adhesion markers implies a partial mesenchymal to epithelial transition.
Conclusions: We conclude that severe hypoxia confers a quiescent phenotype in hMSCs that is reflected by both the transcriptome profile and gene-specific changes of splicosome actions. The results reveal that severe hypoxia imposes markedly different patterns of gene regulation of MSCs compared with more moderate hypoxia. This is the first study to report hypoxia-regulation of AS in stem/progenitor cells and the first molecular genetic characterization of MSC in a hypoxia-induced quiescent immobile state.
Figures




References
-
- Eliasson P, Rehn M, Hammar P, Larsson P, Sirenko O, Flippin LA, Cammenga J, Jönsson JI. Hypoxia mediates low cell-cycle activity and increases the proportion of long-term reconstituting hematopoietic stem cells during in vitro culture. Exp Hematol. 2010;38:301–310. doi: 10.1016/j.exphem.2010.01.005. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

