In silico design and enzymatic synthesis of functional RNA nanoparticles
- PMID: 24758371
- PMCID: PMC4066900
- DOI: 10.1021/ar400329z
In silico design and enzymatic synthesis of functional RNA nanoparticles
Abstract
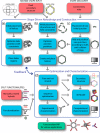
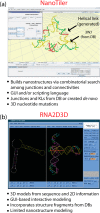
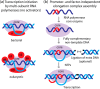
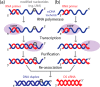
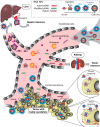
CONSPECTUS: The use of RNAs as scaffolds for biomedical applications has several advantages compared with other existing nanomaterials. These include (i) programmability, (ii) precise control over folding and self-assembly, (iii) natural functionalities as exemplified by ribozymes, riboswitches, RNAi, editing, splicing, and inherent translation and transcription control mechanisms, (iv) biocompatibility, (v) relatively low immune response, and (vi) relatively low cost and ease of production. We have tapped into several of these properties and functionalities to construct RNA-based functional nanoparticles (RNA NPs). In several cases, the structural core and the functional components of the NPs are inherent in the same construct. This permits control over the spatial disposition of the components, intracellular availability, and precise stoichiometry. To enable the generation of RNA NPs, a pipeline is being developed. On one end, it encompasses the rational design and various computational schemes that promote design of the RNA-based nanoconstructs, ultimately producing a set of sequences consisting of RNA or RNA-DNA hybrids, which can assemble into the designed construct. On the other end of the pipeline is an experimental component, which takes the produced sequences and uses them to initialize and characterize their proper assembly and then test the resulting RNA NPs for their function and delivery in cell culture and animal models. An important aspect of this pipeline is the feedback that constantly occurs between the computational and the experimental parts, which synergizes the refinement of both the algorithmic methodologies and the experimental protocols. The utility of this approach is depicted by the several examples described in this Account (nanocubes, nanorings, and RNA-DNA hybrids). Of particular interest, from the computational viewpoint, is that in most cases, first a three-dimensional representation of the assembly is produced, and only then are algorithms applied to generate the sequences that will assemble into the designated three-dimensional construct. This is opposite to the usual practice of predicting RNA structures from a given sequence, that is, the RNA folding problem. To be considered is the generation of sequences that upon assembly have the proper intra- or interstrand interactions (or both). Of particular interest from the experimental point of view is the determination and characterization of the proper thermodynamic, kinetic, functionality, and delivery protocols. Assembly of RNA NPs from individual single-stranded RNAs can be accomplished by one-pot techniques under the proper thermal and buffer conditions or, potentially more interestingly, by the use of various RNA polymerases that can promote the formation of RNA NPs cotransciptionally from specifically designed DNA templates. Also of importance is the delivery of the RNA NPs to the cells of interest in vitro or in vivo. Nonmodified RNAs rapidly degrade in blood serum and have difficulties crossing biological membranes due to their negative charge. These problems can be overcome by using, for example, polycationic lipid-based carriers. Our work involves the use of bolaamphiphiles, which are amphipathic compounds with positively charged hydrophilic head groups at each end connected by a hydrophobic chain. We have correlated results from molecular dynamics computations with various experiments to understand the characteristics of such delivery agents.
Figures







References
-
- Seeman N. C. DNA in a material world. Nature 2003, 421, 427–431. - PubMed
-
- Chen J. H.; Seeman N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633. - PubMed
-
- Han D.; Pal S.; Yang Y.; Jiang S.; Nangreave J.; Liu Y.; Yan H. DNA gridiron nanostructures based on four-arm junctions. Science 2013, 339, 1412–1415. - PubMed
-
- He Y.; Ye T.; Su M.; Zhang C.; Ribbe A. E.; Jiang W.; Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 2008, 452, 198–201. - PubMed
-
- Mao C.; LaBean T. H.; Relf J. H.; Seeman N. C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 2000, 407, 493–496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

