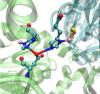
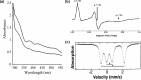
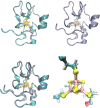
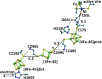
Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers
- PMID: 24758379
- PMCID: PMC4002152
- DOI: 10.1021/cr400479b
Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers
Figures








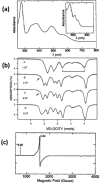



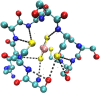







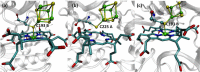




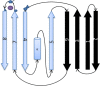

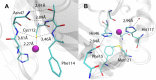
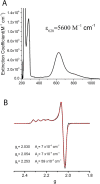

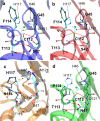
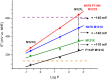
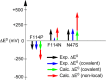





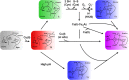




References
-
- Malkin R.; Rabinowitz J. C. Annu. Rev. Biochem. 1967, 36, 113. - PubMed
-
- Adman E. T. Biochim. Biophys. Acta, Rev. Bioenerg. 1979, 549, 107. - PubMed
-
- Brunori M.; Colosimo A.; Silvestrini M. C. Pure Appl. Chem. 1983, 55, 1049.
-
- Thomson A. J. Nachr. Chem., Tech. Lab. 1984, 32, 326.
-
- Chiara S.; Maria; Brunori M. Life Chem. Rep. 1987, 5, 155.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

