Oncogenic Ras induces inflammatory cytokine production by upregulating the squamous cell carcinoma antigens SerpinB3/B4
- PMID: 24759783
- PMCID: PMC4025922
- DOI: 10.1038/ncomms4729
Oncogenic Ras induces inflammatory cytokine production by upregulating the squamous cell carcinoma antigens SerpinB3/B4
Abstract
Mounting evidence indicates that oncogenic Ras can modulate cell autonomous inflammatory cytokine production, although the underlying mechanism remains unclear. Here we show that squamous cell carcinoma antigens 1 and 2 (SCCA1/2), members of the Serpin family of serine/cysteine protease inhibitors, are transcriptionally upregulated by oncogenic Ras via MAPK and the ETS family transcription factor PEA3. Increased SCCA expression leads to inhibition of protein turnover, unfolded protein response, activation of NF-κB and is essential for Ras-mediated cytokine production and tumour growth. Analysis of human colorectal and pancreatic tumour samples reveals a positive correlation between Ras mutation, enhanced SCCA expression and IL-6 expression. These results indicate that SCCA is a Ras-responsive factor that plays an important role in Ras-associated cytokine production and tumorigenesis.
Figures

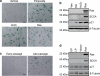
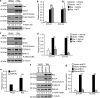

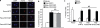



References
-
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. - PubMed
-
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. - PubMed
-
- Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. - PubMed
-
- Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50CA095103/CA/NCI NIH HHS/United States
- P50 CA095103/CA/NCI NIH HHS/United States
- R01GM97355/GM/NIGMS NIH HHS/United States
- R01CA100126/CA/NCI NIH HHS/United States
- R01 GM097355/GM/NIGMS NIH HHS/United States
- U01CA168409/CA/NCI NIH HHS/United States
- R01CA159222/CA/NCI NIH HHS/United States
- T32 CA009176/CA/NCI NIH HHS/United States
- R01CA129536/CA/NCI NIH HHS/United States
- R01 CA129536/CA/NCI NIH HHS/United States
- R01 CA159222/CA/NCI NIH HHS/United States
- R01 CA100126/CA/NCI NIH HHS/United States
- U01 CA168409/CA/NCI NIH HHS/United States
- T32CA009176/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

