Culture-independent evaluation of the appendix and rectum microbiomes in children with and without appendicitis
- PMID: 24759879
- PMCID: PMC3997405
- DOI: 10.1371/journal.pone.0095414
Culture-independent evaluation of the appendix and rectum microbiomes in children with and without appendicitis
Abstract
Purpose: The function of the appendix is largely unknown, but its microbiota likely contributes to function. Alterations in microbiota may contribute to appendicitis, but conventional culture studies have not yielded conclusive information. We conducted a pilot, culture-independent 16S rRNA-based microbiota study of paired appendix and rectal samples.
Methods: We collected appendix and rectal swabs from 21 children undergoing appendectomy, six with normal appendices and fifteen with appendicitis (nine perforated). After DNA extraction, we amplified and sequenced 16S rRNA genes and analyzed sequences using CLoVR. We identified organisms differing in relative abundance using ANOVA (p<0.05) by location (appendix vs. rectum), disease (appendicitis vs. normal), and disease severity (perforated vs. non-perforated).
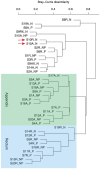
Results: We identified 290 taxa in the study's samples. Three taxa were significantly increased in normal appendices vs. normal rectal samples: Fusibacter (p = 0.009), Selenomonas (p = 0.026), and Peptostreptococcus (p = 0.049). Five taxa were increased in abundance in normal vs. diseased appendices: Paenibacillaceae (p = 0.005), Acidobacteriaceae GP4 (p = 0.019), Pseudonocardinae (p = 0.019), Bergeyella (p = 0.019) and Rhizobium (p = 0.045). Twelve taxa were increased in the appendices of appendicitis patients vs. normal appendix: Peptostreptococcus (p = 0.0003), Bilophila (p = 0.0004), Bulleidia (p = 0.012), Fusobacterium (p = 0.018), Parvimonas (p = 0.003), Mogibacterium (p = 0.012), Aminobacterium (p = 0.019), Proteus (p = 0.028), Actinomycineae (p = 0.028), Anaerovorax (p = 0.041), Anaerofilum (p = 0.045), Porphyromonas (p = 0.010). Five taxa were increased in appendices in patients with perforated vs. nonperforated appendicitis: Bulleidia (p = 0.004), Fusibacter (p = 0.005), Prevotella (p = 0.021), Porphyromonas (p = 0.030), Dialister (p = 0.035). Three taxa were increased in rectum samples of patients with appendicitis compared to the normal patients: Bulleidia (p = 0.034), Dialister (p = 0.003), and Porphyromonas (p = 0.026).
Conclusion: Specific taxa are more abundant in normal appendices compared to the rectum, suggesting that a distinctive appendix microbiota exists. Taxa with altered abundance in diseased and severely diseased (perforated) samples may contribute to appendicitis pathogenesis, and may provide microbial signatures in the rectum useful for guiding both treatment and diagnosis of appendicitis.
Conflict of interest statement
Figures


References
-
- Gebbers JO, Laissue JA (2004) Bacterial translocation in the normal human appendix parallels the development of the local immune system. Ann NY Acad Sci 1029: 337–343. - PubMed
-
- Randal Bollinger R, Barbas AS, Bush EL, Lin SS, Parker W (2007) Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J Theor Biol 249: 826–831. - PubMed
-
- Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL (2004) Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J Immunol 172: 1118–1124. - PubMed
-
- Fitz R (1886) Perforating inflammation of the vermiform appendix. Philadelphia, PA: Wm. J. Dornan, Printer. 74 p.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

