Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins
- PMID: 24760129
- PMCID: PMC4131146
- DOI: 10.1007/s00018-014-1627-y
Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins
Abstract
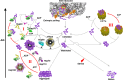
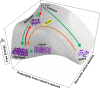
By virtue of their general ability to bind (hold) translocating or unfolding polypeptides otherwise doomed to aggregate, molecular chaperones are commonly dubbed "holdases". Yet, chaperones also carry physiological functions that do not necessitate prevention of aggregation, such as altering the native states of proteins, as in the disassembly of SNARE complexes and clathrin coats. To carry such physiological functions, major members of the Hsp70, Hsp110, Hsp100, and Hsp60/CCT chaperone families act as catalytic unfolding enzymes or unfoldases that drive iterative cycles of protein binding, unfolding/pulling, and release. One unfoldase chaperone may thus successively convert many misfolded or alternatively folded polypeptide substrates into transiently unfolded intermediates, which, once released, can spontaneously refold into low-affinity native products. Whereas during stress, a large excess of non-catalytic chaperones in holding mode may optimally prevent protein aggregation, after the stress, catalytic disaggregases and unfoldases may act as nanomachines that use the energy of ATP hydrolysis to repair proteins with compromised conformations. Thus, holding and catalytic unfolding chaperones can act as primary cellular defenses against the formation of early misfolded and aggregated proteotoxic conformers in order to avert or retard the onset of degenerative protein conformational diseases.
Figures
Similar articles
-
Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides.FEBS Lett. 2013 Jun 27;587(13):1981-7. doi: 10.1016/j.febslet.2013.05.014. Epub 2013 May 16. FEBS Lett. 2013. PMID: 23684649 Review.
-
Disaggregating chaperones: an unfolding story.Curr Protein Pept Sci. 2009 Oct;10(5):432-46. doi: 10.2174/138920309789351930. Curr Protein Pept Sci. 2009. PMID: 19538153 Review.
-
Experimental Milestones in the Discovery of Molecular Chaperones as Polypeptide Unfolding Enzymes.Annu Rev Biochem. 2016 Jun 2;85:715-42. doi: 10.1146/annurev-biochem-060815-014124. Epub 2016 Mar 31. Annu Rev Biochem. 2016. PMID: 27050154 Review.
-
Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones.Front Mol Biosci. 2015 Jun 5;2:29. doi: 10.3389/fmolb.2015.00029. eCollection 2015. Front Mol Biosci. 2015. PMID: 26097841 Free PMC article. Review.
-
Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates.J Biol Chem. 2013 Jul 19;288(29):21399-21411. doi: 10.1074/jbc.M113.479253. Epub 2013 Jun 4. J Biol Chem. 2013. PMID: 23737532 Free PMC article.
Cited by
-
Application of Data-Independent Acquisition Approach to Study the Proteome Change from Early to Later Phases of Tomato Pathogenesis Responses.Int J Mol Sci. 2019 Feb 17;20(4):863. doi: 10.3390/ijms20040863. Int J Mol Sci. 2019. PMID: 30781546 Free PMC article.
-
Modulation of Amyloid States by Molecular Chaperones.Cold Spring Harb Perspect Biol. 2019 Jul 1;11(7):a033969. doi: 10.1101/cshperspect.a033969. Cold Spring Harb Perspect Biol. 2019. PMID: 30755450 Free PMC article. Review.
-
Redefining Molecular Chaperones as Chaotropes.Front Mol Biosci. 2021 Jun 14;8:683132. doi: 10.3389/fmolb.2021.683132. eCollection 2021. Front Mol Biosci. 2021. PMID: 34195228 Free PMC article. Review.
-
Heat shock proteins and hormesis in the diagnosis and treatment of neurodegenerative diseases.Immun Ageing. 2015 Nov 4;12:20. doi: 10.1186/s12979-015-0046-8. eCollection 2015. Immun Ageing. 2015. PMID: 26543490 Free PMC article. Review.
-
Quantitative proteomic analysis to capture the role of heat-accumulated proteins in moss plant acquired thermotolerance.Plant Cell Environ. 2021 Jul;44(7):2117-2133. doi: 10.1111/pce.13975. Epub 2020 Dec 21. Plant Cell Environ. 2021. PMID: 33314263 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

