Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis
- PMID: 24760347
- PMCID: PMC4009603
- DOI: 10.1101/gr.162131.113
Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis
Abstract
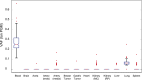
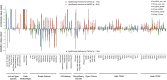
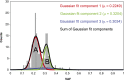
The somatic mutation burden in healthy white blood cells (WBCs) is not well known. Based on deep whole-genome sequencing, we estimate that approximately 450 somatic mutations accumulated in the nonrepetitive genome within the healthy blood compartment of a 115-yr-old woman. The detected mutations appear to have been harmless passenger mutations: They were enriched in noncoding, AT-rich regions that are not evolutionarily conserved, and they were depleted for genomic elements where mutations might have favorable or adverse effects on cellular fitness, such as regions with actively transcribed genes. The distribution of variant allele frequencies of these mutations suggests that the majority of the peripheral white blood cells were offspring of two related hematopoietic stem cell (HSC) clones. Moreover, telomere lengths of the WBCs were significantly shorter than telomere lengths from other tissues. Together, this suggests that the finite lifespan of HSCs, rather than somatic mutation effects, may lead to hematopoietic clonal evolution at extreme ages.
Figures


References
-
- Abkowitz JL, Catlin SN, McCallie MT, Guttorp P 2002. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood 100: 2665–2667 - PubMed
-
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41: 1207–1215 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
