HDAC1 and HDAC2 control the specification of neural crest cells into peripheral glia
- PMID: 24760871
- PMCID: PMC3996228
- DOI: 10.1523/JNEUROSCI.5212-13.2014
HDAC1 and HDAC2 control the specification of neural crest cells into peripheral glia
Abstract
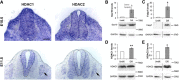
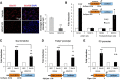
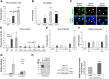
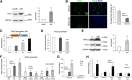
Schwann cells, the myelinating glia of the peripheral nervous system (PNS), originate from multipotent neural crest cells that also give rise to other cells, including neurons, melanocytes, chondrocytes, and smooth muscle cells. The transcription factor Sox10 is required for peripheral glia specification. However, all neural crest cells express Sox10 and the mechanisms directing neural crest cells into a specific lineage are poorly understood. We show here that histone deacetylases 1 and 2 (HDAC1/2) are essential for the specification of neural crest cells into Schwann cell precursors and satellite glia, which express the early determinants of their lineage myelin protein zero (P0) and/or fatty acid binding protein 7 (Fabp7). In neural crest cells, HDAC1/2 induced expression of the transcription factor Pax3 by binding and activating the Pax3 promoter. In turn, Pax3 was required to maintain high Sox10 levels and to trigger expression of Fabp7. In addition, HDAC1/2 were bound to the P0 promoter and activated P0 transcription. Consistently, in vivo genetic deletion of HDAC1/2 in mouse neural crest cells led to strongly decreased Sox10 expression, no detectable Pax3, virtually no satellite glia, and no Schwann cell precursors in dorsal root ganglia and peripheral nerves. Similarly, in vivo ablation of Pax3 in the mouse neural crest resulted in strongly reduced expression of Sox10 and Fabp7. Therefore, by controlling the expression of Pax3 and the concerted action of Pax3 and Sox10 on their target genes, HDAC1/2 direct the specification of neural crest cells into peripheral glia.
Keywords: histone deacetylases; neural crest cells; peripheral glia specification; transcriptional control.
Figures
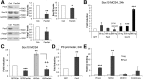

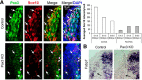
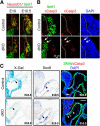
References
-
- Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. - DOI - PMC - PubMed
-
- Auerbach R. Analysis of the developmental effects of a lethal mutation in the house mouse. Journal of Experimental Zoology. 1954;127:305–329. doi: 10.1002/jez.1401270206. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
