Statistical linkage analysis of substitutions in patient-derived sequences of genotype 1a hepatitis C virus nonstructural protein 3 exposes targets for immunogen design
- PMID: 24760894
- PMCID: PMC4054436
- DOI: 10.1128/JVI.03812-13
Statistical linkage analysis of substitutions in patient-derived sequences of genotype 1a hepatitis C virus nonstructural protein 3 exposes targets for immunogen design
Abstract
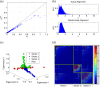
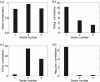
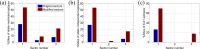
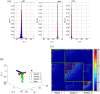
Chronic hepatitis C virus (HCV) infection is one of the leading causes of liver failure and liver cancer, affecting around 3% of the world's population. The extreme sequence variability of the virus resulting from error-prone replication has thwarted the discovery of a universal prophylactic vaccine. It is known that vigorous and multispecific cellular immune responses, involving both helper CD4(+) and cytotoxic CD8(+) T cells, are associated with the spontaneous clearance of acute HCV infection. Escape mutations in viral epitopes can, however, abrogate protective T-cell responses, leading to viral persistence and associated pathologies. Despite the propensity of the virus to mutate, there might still exist substitutions that incur a fitness cost. In this paper, we identify groups of coevolving residues within HCV nonstructural protein 3 (NS3) by analyzing diverse sequences of this protein using ideas from random matrix theory and associated methods. Our analyses indicate that one of these groups comprises a large percentage of residues for which HCV appears to resist multiple simultaneous substitutions. Targeting multiple residues in this group through vaccine-induced immune responses should either lead to viral recognition or elicit escape substitutions that compromise viral fitness. Our predictions are supported by published clinical data, which suggested that immune genotypes associated with spontaneous clearance of HCV preferentially recognized and targeted this vulnerable group of residues. Moreover, mapping the sites of this group onto the available protein structure provided insight into its functional significance. An epitope-based immunogen is proposed as an alternative to the NS3 epitopes in the peptide-based vaccine IC41.
Importance: Despite much experimental work on HCV, a thorough statistical study of the HCV sequences for the purpose of immunogen design was missing in the literature. Such a study is vital to identify epistatic couplings among residues that can provide useful insights for designing a potent vaccine. In this work, ideas from random matrix theory were applied to characterize the statistics of substitutions within the diverse publicly available sequences of the genotype 1a HCV NS3 protein, leading to a group of sites for which HCV appears to resist simultaneous substitutions possibly due to deleterious effect on viral fitness. Our analysis leads to completely novel immunogen designs for HCV. In addition, the NS3 epitopes used in the recently proposed peptide-based vaccine IC41 were analyzed in the context of our framework. Our analysis predicts that alternative NS3 epitopes may be worth exploring as they might be more efficacious.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- World Health Organization. 2013. Factsheet, hepatitis C. World Health Organization, Geneva, Switzerland
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

