Polyglutamine Disease Modeling: Epitope Based Screen for Homologous Recombination using CRISPR/Cas9 System
- PMID: 24761311
- PMCID: PMC3994193
- DOI: 10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a
Polyglutamine Disease Modeling: Epitope Based Screen for Homologous Recombination using CRISPR/Cas9 System
Abstract
We have previously reported the genetic correction of Huntington's disease (HD) patient-derived induced pluripotent stem cells using traditional homologous recombination (HR) approaches. To extend this work, we have adopted a CRISPR-based genome editing approach to improve the efficiency of recombination in order to generate allelic isogenic HD models in human cells. Incorporation of a rapid antibody-based screening approach to measure recombination provides a powerful method to determine relative efficiency of genome editing for modeling polyglutamine diseases or understanding factors that modulate CRISPR/Cas9 HR.
Figures
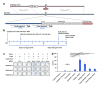
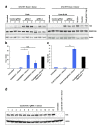
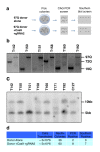

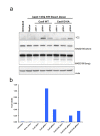
Similar articles
-
A Human Induced Pluripotent Stem Cell-Derived Isogenic Model of Huntington's Disease Based on Neuronal Cells Has Several Relevant Phenotypic Abnormalities.J Pers Med. 2020 Nov 9;10(4):215. doi: 10.3390/jpm10040215. J Pers Med. 2020. PMID: 33182269 Free PMC article.
-
Generation of New Isogenic Models of Huntington's Disease Using CRISPR-Cas9 Technology.Int J Mol Sci. 2020 Mar 8;21(5):1854. doi: 10.3390/ijms21051854. Int J Mol Sci. 2020. PMID: 32182692 Free PMC article.
-
Footprint-free gene mutation correction in induced pluripotent stem cell (iPSC) derived from recessive dystrophic epidermolysis bullosa (RDEB) using the CRISPR/Cas9 and piggyBac transposon system.J Dermatol Sci. 2020 Jun;98(3):163-172. doi: 10.1016/j.jdermsci.2020.04.004. Epub 2020 Apr 24. J Dermatol Sci. 2020. PMID: 32376152
-
Modern Genome Editing Technologies in Huntington's Disease Research.J Huntingtons Dis. 2017;6(1):19-31. doi: 10.3233/JHD-160222. J Huntingtons Dis. 2017. PMID: 28128770 Free PMC article. Review.
-
CRISPR/Cas9 Gene Editing: From Basic Mechanisms to Improved Strategies for Enhanced Genome Engineering In Vivo.Curr Gene Ther. 2017;17(4):263-274. doi: 10.2174/1566523217666171122094629. Curr Gene Ther. 2017. PMID: 29173169 Review.
Cited by
-
The prospects of CRISPR-based genome engineering in the treatment of neurodegenerative disorders.Ther Adv Neurol Disord. 2017 Nov 15;11:1756285617741837. doi: 10.1177/1756285617741837. eCollection 2018. Ther Adv Neurol Disord. 2017. PMID: 29399048 Free PMC article. Review.
-
Precise Excision of the CAG Tract from the Huntingtin Gene by Cas9 Nickases.Front Neurosci. 2018 Feb 26;12:75. doi: 10.3389/fnins.2018.00075. eCollection 2018. Front Neurosci. 2018. PMID: 29535594 Free PMC article.
-
iPSC-based drug screening for Huntington's disease.Brain Res. 2016 May 1;1638(Pt A):42-56. doi: 10.1016/j.brainres.2015.09.020. Epub 2015 Sep 30. Brain Res. 2016. PMID: 26428226 Free PMC article. Review.
-
Genome Engineering with TALE and CRISPR Systems in Neuroscience.Front Genet. 2016 Apr 6;7:47. doi: 10.3389/fgene.2016.00047. eCollection 2016. Front Genet. 2016. PMID: 27092173 Free PMC article. Review.
-
Cell-Autonomous and Non-cell-Autonomous Pathogenic Mechanisms in Huntington's Disease: Insights from In Vitro and In Vivo Models.Neurotherapeutics. 2019 Oct;16(4):957-978. doi: 10.1007/s13311-019-00782-9. Neurotherapeutics. 2019. PMID: 31529216 Free PMC article. Review.
References
-
- Yang, L., Mali, P., Kim-Kiselak, C. & Church, G. CRISPR-Cas-Mediated Targeted Genome Editing in Human Cells. Methods Mol Biol 1114, 245-267, doi:10.1007/978-1-62703-761-7_16 (2014). - PubMed
-
- Bassett, A. R. & Liu, J. L. CRISPR/Cas9 and Genome Editing in Drosophila. Journal of genetics and genomics = Yi chuan xue bao 41, 7-19, doi:10.1016/j.jgg.2013.12.004 (2014). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

