Mesenchymal gene program-expressing ovarian cancer spheroids exhibit enhanced mesothelial clearance
- PMID: 24762435
- PMCID: PMC4038562
- DOI: 10.1172/JCI69815
Mesenchymal gene program-expressing ovarian cancer spheroids exhibit enhanced mesothelial clearance
Abstract
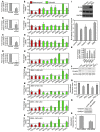
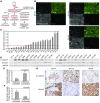
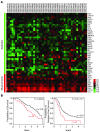
Metastatic dissemination of ovarian tumors involves the invasion of tumor cell clusters into the mesothelial cell lining of peritoneal cavity organs; however, the tumor-specific factors that allow ovarian cancer cells to spread are unclear. We used an in vitro assay that models the initial step of ovarian cancer metastasis, clearance of the mesothelial cell layer, to examine the clearance ability of a large panel of both established and primary ovarian tumor cells. Comparison of the gene and protein expression profiles of clearance-competent and clearance-incompetent cells revealed that mesenchymal genes are enriched in tumor populations that display strong clearance activity, while epithelial genes are enriched in those with weak or undetectable activity. Overexpression of transcription factors SNAI1, TWIST1, and ZEB1, which regulate the epithelial-to-mesenchymal transition (EMT), promoted mesothelial clearance in cell lines with weak activity, while knockdown of the EMT-regulatory transcription factors TWIST1 and ZEB1 attenuated mesothelial clearance in ovarian cancer cell lines with strong activity. These findings provide important insights into the mechanisms associated with metastatic progression of ovarian cancer and suggest that inhibiting pathways that drive mesenchymal programs may suppress tumor cell invasion of peritoneal tissues.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

