Comparison of filtering methods for the modeling and retrospective forecasting of influenza epidemics
- PMID: 24762780
- PMCID: PMC3998879
- DOI: 10.1371/journal.pcbi.1003583
Comparison of filtering methods for the modeling and retrospective forecasting of influenza epidemics
Abstract
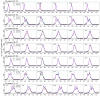
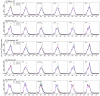
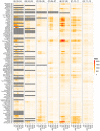
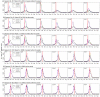
A variety of filtering methods enable the recursive estimation of system state variables and inference of model parameters. These methods have found application in a range of disciplines and settings, including engineering design and forecasting, and, over the last two decades, have been applied to infectious disease epidemiology. For any system of interest, the ideal filter depends on the nonlinearity and complexity of the model to which it is applied, the quality and abundance of observations being entrained, and the ultimate application (e.g. forecast, parameter estimation, etc.). Here, we compare the performance of six state-of-the-art filter methods when used to model and forecast influenza activity. Three particle filters--a basic particle filter (PF) with resampling and regularization, maximum likelihood estimation via iterated filtering (MIF), and particle Markov chain Monte Carlo (pMCMC)--and three ensemble filters--the ensemble Kalman filter (EnKF), the ensemble adjustment Kalman filter (EAKF), and the rank histogram filter (RHF)--were used in conjunction with a humidity-forced susceptible-infectious-recovered-susceptible (SIRS) model and weekly estimates of influenza incidence. The modeling frameworks, first validated with synthetic influenza epidemic data, were then applied to fit and retrospectively forecast the historical incidence time series of seven influenza epidemics during 2003-2012, for 115 cities in the United States. Results suggest that when using the SIRS model the ensemble filters and the basic PF are more capable of faithfully recreating historical influenza incidence time series, while the MIF and pMCMC do not perform as well for multimodal outbreaks. For forecast of the week with the highest influenza activity, the accuracies of the six model-filter frameworks are comparable; the three particle filters perform slightly better predicting peaks 1-5 weeks in the future; the ensemble filters are more accurate predicting peaks in the past.
Conflict of interest statement
The authors have declared that no competing interests exist.
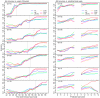
Figures


References
-
- Molinari N-AM, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. (2007) The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine 25: 5086–5096. - PubMed
-
- Skvortsov A, Ristic B (2012) Monitoring and prediction of an epidemic outbreak using syndromic observations. Mathematical Biosciences 240: 12–19. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

