Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy
- PMID: 24762976
- PMCID: PMC4143415
- DOI: 10.4161/epi.28945
Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy
Erratum in
-
Corrigendum.Epigenetics. 2015;10(10):994. doi: 10.1080/15592294.2015.1078144. Epigenetics. 2015. PMID: 26453355 Free PMC article. No abstract available.
Abstract
Food allergy is mediated by a combination of genetic and environmental risk factors, potentially mediated by epigenetic mechanisms. CD4+ T-cells are key drivers of the allergic response, and may therefore harbor epigenetic variation in association with the disease phenotype. Here we retrospectively examined genome-wide DNA methylation profiles (~450,000 CpGs) from CD4+ T-cells on a birth cohort of 12 children with IgE-mediated food allergy diagnosed at 12-months, and 12 non-allergic controls. DNA samples were available at two time points, birth and 12-months.
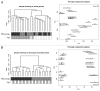
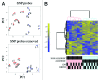
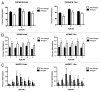
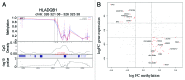
Case: control comparisons of CD4+ methylation profiles identified 179 differentially methylated probes (DMP) at 12-months and 136 DMP at birth (FDR-adjusted P value<0.05, delta β>0.1). Approximately 30% of DMPs were coincident with previously annotated SNPs. A total of 92 [corrected] allergy-associated non-SNP DMPs were present at birth when individuals were initially disease-free, potentially implicating these loci in the causal pathway. Pathway analysis of differentially methylated genes identified several MAP kinase signaling molecules. Mass spectrometry was used to validate 15 CpG sites at 3 candidate genes. Combined analysis of differential methylation with gene expression profiles revealed gene expression differences at some but not all allergy associated differentially methylated genes. Thus, dysregulation of DNA methylation at MAPK signaling-associated genes during early CD4+ T-cell development may contribute to suboptimal T-lymphocyte responses in early childhood associated with the development of food allergy.
Keywords: EWAS; Infinium 450k; allergic disease; food allergy; in utero programming; metastable epialleles.
Figures

References
-
- Osborne NJ, Koplin JJ, Martin PE, Gurrin LC, Lowe AJ, Matheson MC, Ponsonby A-L, Wake M, Tang MLK, Dharmage SC, et al. HealthNuts Investigators Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol. 2011;127:668–, e1-2. doi: 10.1016/j.jaci.2011.01.039. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
