A novel highly divergent protein family identified from a viviparous insect by RNA-seq analysis: a potential target for tsetse fly-specific abortifacients
- PMID: 24763277
- PMCID: PMC3998918
- DOI: 10.1371/journal.pgen.1003874
A novel highly divergent protein family identified from a viviparous insect by RNA-seq analysis: a potential target for tsetse fly-specific abortifacients
Abstract
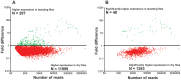
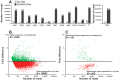
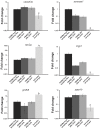
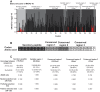
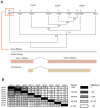
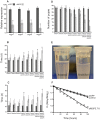
In tsetse flies, nutrients for intrauterine larval development are synthesized by the modified accessory gland (milk gland) and provided in mother's milk during lactation. Interference with at least two milk proteins has been shown to extend larval development and reduce fecundity. The goal of this study was to perform a comprehensive characterization of tsetse milk proteins using lactation-specific transcriptome/milk proteome analyses and to define functional role(s) for the milk proteins during lactation. Differential analysis of RNA-seq data from lactating and dry (non-lactating) females revealed enrichment of transcripts coding for protein synthesis machinery, lipid metabolism and secretory proteins during lactation. Among the genes induced during lactation were those encoding the previously identified milk proteins (milk gland proteins 1-3, transferrin and acid sphingomyelinase 1) and seven new genes (mgp4-10). The genes encoding mgp2-10 are organized on a 40 kb syntenic block in the tsetse genome, have similar exon-intron arrangements, and share regions of amino acid sequence similarity. Expression of mgp2-10 is female-specific and high during milk secretion. While knockdown of a single mgp failed to reduce fecundity, simultaneous knockdown of multiple variants reduced milk protein levels and lowered fecundity. The genomic localization, gene structure similarities, and functional redundancy of MGP2-10 suggest that they constitute a novel highly divergent protein family. Our data indicates that MGP2-10 function both as the primary amino acid resource for the developing larva and in the maintenance of milk homeostasis, similar to the function of the mammalian casein family of milk proteins. This study underscores the dynamic nature of the lactation cycle and identifies a novel family of lactation-specific proteins, unique to Glossina sp., that are essential to larval development. The specificity of MGP2-10 to tsetse and their critical role during lactation suggests that these proteins may be an excellent target for tsetse-specific population control approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Tobe SS, Langley PA (1978) Reproductive physiology of Glossina . Ann Rev Entomol 23: 283–307. - PubMed
-
- Cmelik SHW, Bursell E, Slack E (1969) Composition of the gut contents of thrid-instar tsetse larvae (Glossina morsitans Westwood). Comp Biochem Physiol 29: 447–453.
-
- Ejezie GC, Davey KG (1976) Some effects of allatectomy in female tsetse, Glossina austeni . J Insect Physiol 22: 1743–1749. - PubMed
-
- Denlinger DL (1975) Insect hormones as tsetse abortifacients. Nature 253: 347–348. - PubMed
-
- Denlinger DL, Ma W-C (1974) Dynamics of the pregnancy cycle in the tsetse Glossina morsitans . J Insect Physiol 20: 1015–1026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

