FRA2A is a CGG repeat expansion associated with silencing of AFF3
- PMID: 24763282
- PMCID: PMC3998887
- DOI: 10.1371/journal.pgen.1004242
FRA2A is a CGG repeat expansion associated with silencing of AFF3
Abstract
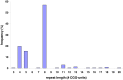
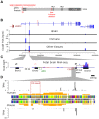
Folate-sensitive fragile sites (FSFS) are a rare cytogenetically visible subset of dynamic mutations. Of the eight molecularly characterized FSFS, four are associated with intellectual disability (ID). Cytogenetic expression results from CGG tri-nucleotide-repeat expansion mutation associated with local CpG hypermethylation and transcriptional silencing. The best studied is the FRAXA site in the FMR1 gene, where large expansions cause fragile X syndrome, the most common inherited ID syndrome. Here we studied three families with FRA2A expression at 2q11 associated with a wide spectrum of neurodevelopmental phenotypes. We identified a polymorphic CGG repeat in a conserved, brain-active alternative promoter of the AFF3 gene, an autosomal homolog of the X-linked AFF2/FMR2 gene: Expansion of the AFF2 CGG repeat causes FRAXE ID. We found that FRA2A-expressing individuals have mosaic expansions of the AFF3 CGG repeat in the range of several hundred repeat units. Moreover, bisulfite sequencing and pyrosequencing both suggest AFF3 promoter hypermethylation. cSNP-analysis demonstrates monoallelic expression of the AFF3 gene in FRA2A carriers thus predicting that FRA2A expression results in functional haploinsufficiency for AFF3 at least in a subset of tissues. By whole-mount in situ hybridization the mouse AFF3 ortholog shows strong regional expression in the developing brain, somites and limb buds in 9.5-12.5dpc mouse embryos. Our data suggest that there may be an association between FRA2A and a delay in the acquisition of motor and language skills in the families studied here. However, additional cases are required to firmly establish a causal relationship.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

