Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men
- PMID: 24763370
- PMCID: PMC4992075
- DOI: 10.1038/ismej.2014.63
Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men
Abstract
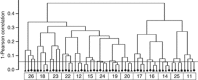
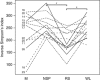
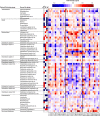
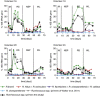
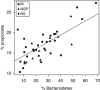
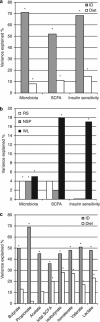
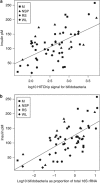
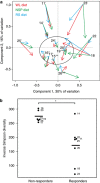
There is growing interest in understanding how diet affects the intestinal microbiota, including its possible associations with systemic diseases such as metabolic syndrome. Here we report a comprehensive and deep microbiota analysis of 14 obese males consuming fully controlled diets supplemented with resistant starch (RS) or non-starch polysaccharides (NSPs) and a weight-loss (WL) diet. We analyzed the composition, diversity and dynamics of the fecal microbiota on each dietary regime by phylogenetic microarray and quantitative PCR (qPCR) analysis. In addition, we analyzed fecal short chain fatty acids (SCFAs) as a proxy of colonic fermentation, and indices of insulin sensitivity from blood samples. The diet explained around 10% of the total variance in microbiota composition, which was substantially less than the inter-individual variance. Yet, each of the study diets induced clear and distinct changes in the microbiota. Multiple Ruminococcaceae phylotypes increased on the RS diet, whereas mostly Lachnospiraceae phylotypes increased on the NSP diet. Bifidobacteria decreased significantly on the WL diet. The RS diet decreased the diversity of the microbiota significantly. The total 16S ribosomal RNA gene signal estimated by qPCR correlated positively with the three major SCFAs, while the amount of propionate specifically correlated with the Bacteroidetes. The dietary responsiveness of the individual's microbiota varied substantially and associated inversely with its diversity, suggesting that individuals can be stratified into responders and non-responders based on the features of their intestinal microbiota.
Figures

References
-
- Bang JW, Crockford DJ, Holmes E, Pazos F, Sternberg MJ, Muggleton SH et al. (2008). Integrative top-down system metabolic modeling in experimental disease states via data-driven Bayesian methods. J Proteome Res 7: 497–503. - PubMed
-
- Becker RA, Chambers JM, Wilks AR. (1988) The new S language: a programming environment for data analysis and graphics, Wadsworth and Brooks. Cole: Pacific Grove.
-
- Burcelin R, Garidou L, Pomié C. (2012). Immuno-microbiota cross and talk: the new paradigm of metabolic diseases. Sem Immunol 24: 67–74. - PubMed
-
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM et al. (2007). Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50: 2374–2383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

