A chloroplast retrograde signal regulates nuclear alternative splicing
- PMID: 24763593
- PMCID: PMC4382720
- DOI: 10.1126/science.1250322
A chloroplast retrograde signal regulates nuclear alternative splicing
Abstract
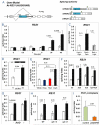
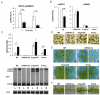
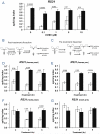
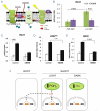
Light is a source of energy and also a regulator of plant physiological adaptations. We show here that light/dark conditions affect alternative splicing of a subset of Arabidopsis genes preferentially encoding proteins involved in RNA processing. The effect requires functional chloroplasts and is also observed in roots when the communication with the photosynthetic tissues is not interrupted, suggesting that a signaling molecule travels through the plant. Using photosynthetic electron transfer inhibitors with different mechanisms of action, we deduce that the reduced pool of plastoquinones initiates a chloroplast retrograde signaling that regulates nuclear alternative splicing and is necessary for proper plant responses to varying light conditions.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

