Nonenzymatic rubylation and ubiquitination of proteins for structural and functional studies
- PMID: 24764216
- PMCID: PMC4128492
- DOI: 10.1002/anie.201402642
Nonenzymatic rubylation and ubiquitination of proteins for structural and functional studies
Abstract
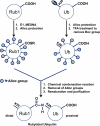
Uncovering the mechanisms that allow conjugates of ubiquitin (Ub) and/or Ub-like (UBL) proteins such as Rub1 to serve as distinct molecular signals requires the ability to make them with native connectivity and defined length and linkage composition. A novel, effective, and affordable strategy for controlled chemical assembly of fully natural UBL-Ub, Ub-UBL, and UBL-UBL conjugates from recombinant monomers is presented. Rubylation of Ub and Rub1 and ubiquitination of Rub1 was achieved without E2/E3 enzymes. New residue-specific information was obtained on the interdomain contacts in naturally-occurring K48-linked Rub1-Ub and Ub-Rub1, and K29-linked Rub1-Ub heterodimers, and their recognition by a K48-linkage-specific Ub receptor. The disassembly of these heterodimers by major deubiquitinating enzymes was examined and it was discovered that some deubiquitinases also possess derubylase activity. This unexpected result suggests possible crosstalk between Ub and Rub1/Nedd8 signaling pathways.
Keywords: deubiquitinases; nonenzymatic assembly; protein modifications; rubylation; ubiquitination.
© 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

Similar articles
-
Strategies to Trap Enzyme-Substrate Complexes that Mimic Michaelis Intermediates During E3-Mediated Ubiquitin-Like Protein Ligation.Methods Mol Biol. 2018;1844:169-196. doi: 10.1007/978-1-4939-8706-1_12. Methods Mol Biol. 2018. PMID: 30242710 Free PMC article.
-
Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system.Mol Cell Proteomics. 2012 Dec;11(12):1595-611. doi: 10.1074/mcp.M112.022467. Epub 2012 Oct 26. Mol Cell Proteomics. 2012. PMID: 23105008 Free PMC article.
-
Profiling DUBs and Ubl-specific proteases with activity-based probes.Methods Enzymol. 2019;618:357-387. doi: 10.1016/bs.mie.2018.12.037. Epub 2019 Feb 14. Methods Enzymol. 2019. PMID: 30850060 Free PMC article.
-
Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways.Curr Opin Struct Biol. 2007 Dec;17(6):726-35. doi: 10.1016/j.sbi.2007.08.018. Epub 2007 Oct 4. Curr Opin Struct Biol. 2007. PMID: 17919899 Free PMC article. Review.
-
Structural and functional insights to ubiquitin-like protein conjugation.Annu Rev Biophys. 2014;43:357-79. doi: 10.1146/annurev-biophys-051013-022958. Annu Rev Biophys. 2014. PMID: 24773014 Free PMC article. Review.
Cited by
-
Hybrid Chains: A Collaboration of Ubiquitin and Ubiquitin-Like Modifiers Introducing Cross-Functionality to the Ubiquitin Code.Front Chem. 2020 Jan 22;7:931. doi: 10.3389/fchem.2019.00931. eCollection 2019. Front Chem. 2020. PMID: 32039151 Free PMC article. Review.
-
Ubiquitination of the Dishevelled DIX domain blocks its head-to-tail polymerization.Nat Commun. 2015 Apr 24;6:6718. doi: 10.1038/ncomms7718. Nat Commun. 2015. PMID: 25907794 Free PMC article.
-
Chemical ubiquitination for decrypting a cellular code.Biochem J. 2016 May 15;473(10):1297-314. doi: 10.1042/BJ20151195. Biochem J. 2016. PMID: 27208213 Free PMC article. Review.
-
Hydrophobic Patch of Ubiquitin is Important for its Optimal Activation by Ubiquitin Activating Enzyme E1.Anal Chem. 2017 Aug 1;89(15):7852-7860. doi: 10.1021/acs.analchem.6b04194. Epub 2017 Jul 20. Anal Chem. 2017. PMID: 28686836 Free PMC article.
-
Genetic Code Expansion Approaches to Decipher the Ubiquitin Code.Chem Rev. 2024 Oct 23;124(20):11544-11584. doi: 10.1021/acs.chemrev.4c00375. Epub 2024 Sep 23. Chem Rev. 2024. PMID: 39311880 Free PMC article. Review.
References
-
- Marmor MD, Yarden Y. Oncogene. 2004;23:2057–2070. - PubMed
- Jackson SP, Durocher D. Mol Cell. 2013;49:795–807. - PubMed
- Ulrich HD, Walden H. Nat Rev Mol Cell Biol. 2010;11:479–489. - PubMed
- Finley D, Bartel B, Varshavsky A. Nature. 1989;338:394–401. - PubMed
- Teixeira LK, Reed SI. Annu Rev Biochem. 2013 - PubMed
- Loureiro J, Ploegh HL. Adv Immunol. 2006;92:225–305. - PMC - PubMed
-
- Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Oncogene. 1999;18:6829–6834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

