Expression cloning and regulation of steroid 5 alpha-reductase, an enzyme essential for male sexual differentiation
- PMID: 2476440
- PMCID: PMC4448950
Expression cloning and regulation of steroid 5 alpha-reductase, an enzyme essential for male sexual differentiation
Abstract
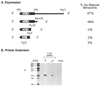
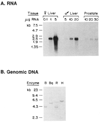
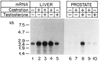
The conversion of testosterone into the more potent androgen, dihydrotestosterone, catalyzed by the enzyme steroid 5 alpha-reductase, is required for the differentiation of male external genitalia. Here, we report the isolation of cDNA clones encoding the rat steroid 5 alpha-reductase using expression cloning in Xenopus oocytes. DNA sequence analysis demonstrates that the liver and ventral prostate forms of steroid 5 alpha-reductases are identical hydrophobic proteins of 29 kDa. The amount of steroid 5 alpha-reductase mRNA in liver increased in response to castration, but remained unchanged in the prostate. Testosterone administration to castrates induced expression of mRNA in the prostate but had no effect on liver. The data suggest that the steroid 5 alpha-reductase gene is differentially regulated by testosterone in androgen-responsive versus non-responsive tissues.
Figures





References
-
- Wilson JD. Annu. Rev. Physiol. 1978;40:279–306. - PubMed
-
- Wilson JD. Handb. Physiol. 1975;5:491–508.
-
- Walsh PC, Madden JD, Harrod MJ, Goldstein JL, MacDonald PC, Wilson JD. N. Engl. J. Med. 1974;291:944–949. - PubMed
-
- Imperato-McGinley J, Guerrero L, Gautier T, Peterson RE. Science. 1974;186:1213–1217. - PubMed
-
- Moore RJ, Wilson JD. Endocrinology. 1973;93:581–592. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

