Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors
- PMID: 24766805
- PMCID: PMC4060823
- DOI: 10.1016/j.cell.2014.04.006
Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors
Erratum in
- Cell. 2014 Jul 3;158(1):226
Abstract
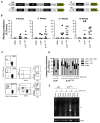
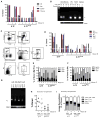
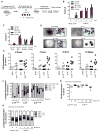
Hematopoietic stem cells (HSCs) sustain blood formation throughout life and are the functional units of bone marrow transplantation. We show that transient expression of six transcription factors Run1t1, Hlf, Lmo2, Prdm5, Pbx1, and Zfp37 imparts multilineage transplantation potential onto otherwise committed lymphoid and myeloid progenitors and myeloid effector cells. Inclusion of Mycn and Meis1 and use of polycistronic viruses increase reprogramming efficacy. The reprogrammed cells, designated induced-HSCs (iHSCs), possess clonal multilineage differentiation potential, reconstitute stem/progenitor compartments, and are serially transplantable. Single-cell analysis revealed that iHSCs derived under optimal conditions exhibit a gene expression profile that is highly similar to endogenous HSCs. These findings demonstrate that expression of a set of defined factors is sufficient to activate the gene networks governing HSC functional identity in committed blood cells. Our results raise the prospect that blood cell reprogramming may be a strategy for derivation of transplantable stem cells for clinical application.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Induced hematopoietic stem cells: unlocking restrictions in lineage potential and self-renewal.Cell Stem Cell. 2014 May 1;14(5):555-6. doi: 10.1016/j.stem.2014.04.008. Cell Stem Cell. 2014. PMID: 24792111
-
'From blood to blood': de-differentiation of hematopoietic progenitors to stem cells.EMBO J. 2014 Jul 17;33(14):1511-3. doi: 10.15252/embj.201488980. Epub 2014 Jun 6. EMBO J. 2014. PMID: 24907133 Free PMC article.
References
-
- Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci U S A. 1990;87:7988–7992. - PMC - PubMed
-
- Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

