Identification of a circadian output circuit for rest:activity rhythms in Drosophila
- PMID: 24766812
- PMCID: PMC4003459
- DOI: 10.1016/j.cell.2014.02.024
Identification of a circadian output circuit for rest:activity rhythms in Drosophila
Abstract
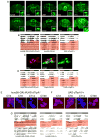
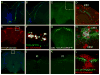
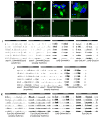
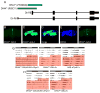
Though much is known about the cellular and molecular components of the circadian clock, output pathways that couple clock cells to overt behaviors have not been identified. We conducted a screen for circadian-relevant neurons in the Drosophila brain and report here that cells of the pars intercerebralis (PI), a functional homolog of the mammalian hypothalamus, comprise an important component of the circadian output pathway for rest:activity rhythms. GFP reconstitution across synaptic partners (GRASP) analysis demonstrates that PI cells are connected to the clock through a polysynaptic circuit extending from pacemaker cells to PI neurons. Molecular profiling of relevant PI cells identified the corticotropin-releasing factor (CRF) homolog, DH44, as a circadian output molecule that is specifically expressed by PI neurons and is required for normal rest:activity rhythms. Notably, selective activation or ablation of just six DH44+ PI cells causes arrhythmicity. These findings delineate a circuit through which clock cells can modulate locomotor rhythms.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
References
-
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

