Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development
- PMID: 24766814
- PMCID: PMC4045247
- DOI: 10.1016/j.cell.2014.04.005
Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development
Abstract
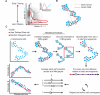
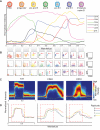
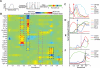
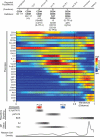
Tissue regeneration is an orchestrated progression of cells from an immature state to a mature one, conventionally represented as distinctive cell subsets. A continuum of transitional cell states exists between these discrete stages. We combine the depth of single-cell mass cytometry and an algorithm developed to leverage this continuum by aligning single cells of a given lineage onto a unified trajectory that accurately predicts the developmental path de novo. Applied to human B cell lymphopoiesis, the algorithm (termed Wanderlust) constructed trajectories spanning from hematopoietic stem cells through to naive B cells. This trajectory revealed nascent fractions of B cell progenitors and aligned them with developmentally cued regulatory signaling including IL-7/STAT5 and cellular events such as immunoglobulin rearrangement, highlighting checkpoints across which regulatory signals are rewired paralleling changes in cellular state. This study provides a comprehensive analysis of human B lymphopoiesis, laying a foundation to apply this approach to other tissues and "corrupted" developmental processes including cancer.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Cobaleda C, Sanchez-García I. B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. BioEssays. 2009;31:600–609. - PubMed
-
- Corfe SA, Paige CJ. The many roles of IL-7 in B cell development; Mediator of survival, proliferation and differentiation. Semin Immunol. 2012;24:198–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA164729/CA/NCI NIH HHS/United States
- HHSF223201210194C/PHS HHS/United States
- U54CA121852-01A1/CA/NCI NIH HHS/United States
- N01-HV-00242/HV/NHLBI NIH HHS/United States
- 41000411217/PHS HHS/United States
- U54CA149145/CA/NCI NIH HHS/United States
- 152175.5041015.0412/PHS HHS/United States
- UL1 RR025744/RR/NCRR NIH HHS/United States
- UL1 TR001085/TR/NCATS NIH HHS/United States
- 5U54CA143907/CA/NCI NIH HHS/United States
- U54 CA149145/CA/NCI NIH HHS/United States
- HHSN272200700038C/AI/NIAID NIH HHS/United States
- DP2-OD002414-01/OD/NIH HHS/United States
- U24 CA224309/CA/NCI NIH HHS/United States
- 5-24927/PHS HHS/United States
- R01 CA130826/CA/NCI NIH HHS/United States
- P01 CA034233-22A1/CA/NCI NIH HHS/United States
- U54 CA143907/CA/NCI NIH HHS/United States
- PN2 EY018228/EY/NEI NIH HHS/United States
- S10 SIG S1044027582-01/PHS HHS/United States
- P01 CA034233/CA/NCI NIH HHS/United States
- K99GM104148-01/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- DP2 OD002414/OD/NIH HHS/United States
- S10 RR027582/RR/NCRR NIH HHS/United States
- U19 AI100627/AI/NIAID NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- 1R01CA130826/CA/NCI NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- K99 GM104148/GM/NIGMS NIH HHS/United States
- R00 GM104148/GM/NIGMS NIH HHS/United States
- PN2EY018228/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

