Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies
- PMID: 24767177
- PMCID: PMC4032163
- DOI: 10.1186/1742-4690-11-33
Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies
Abstract
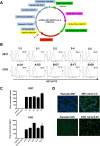
Background: Recombinant soluble, cleaved HIV-1 envelope glycoprotein SOSIP.664 gp140 trimers based on the subtype A BG505 sequence are being studied structurally and tested as immunogens in animals. For these trimers to become a vaccine candidate for human trials, they would need to be made in appropriate amounts at an acceptable quality. Accomplishing such tasks by transient transfection is likely to be challenging. The traditional way to express recombinant proteins in large amounts is via a permanent cell line, usually of mammalian origin. Making cell lines that produce BG505 SOSIP.664 trimers requires the co-expression of the Furin protease to ensure that the cleavage site between the gp120 and gp41 subunits is fully utilized.
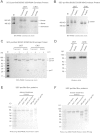
Results: We designed a vector capable of expressing Env and Furin, and used it to create Stable 293 T and CHO Flp-In™ cell lines through site-specific recombination. Both lines produce high quality, cleaved trimers at yields of up to 12-15 mg per 1 × 109 cells. Trimer expression at such levels was maintained for up to 30 days (10 passages) after initial seeding and was consistently superior to what could be achieved by transient transfection. Electron microscopy studies confirm that the purified trimers have the same native-like appearance as those derived by transient transfection and used to generate high-resolution structures. They also have appropriate antigenic properties, including the presentation of the quaternary epitope for the broadly neutralizing antibody PGT145.
Conclusions: The BG505 SOSIP.664 trimer-expressing cell lines yield proteins of an appropriate quality for structural studies and animal immunogenicity experiments. The methodology is suitable for making similar lines under Good Manufacturing Practice conditions, to produce trimers for human clinical trials. Moreover, any env gene can be incorporated into this vector system, allowing the manufacture of SOSIP trimers from multiple genotypes, either by transient transfection or from stable cell lines.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

