Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis
- PMID: 24767259
- PMCID: PMC4051827
- DOI: 10.1016/j.dnarep.2014.03.024
Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis
Abstract
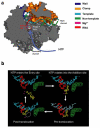
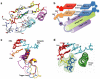
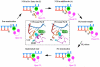
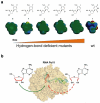
Maintaining high transcriptional fidelity is essential for life. Some DNA lesions lead to significant changes in transcriptional fidelity. In this review, we will summarize recent progress towards understanding the molecular basis of RNA polymerase II (Pol II) transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis. In particular, we will focus on the three key checkpoint steps of controlling Pol II transcriptional fidelity: insertion (specific nucleotide selection and incorporation), extension (differentiation of RNA transcript extension of a matched over mismatched 3'-RNA terminus), and proofreading (preferential removal of misincorporated nucleotides from the 3'-RNA end). We will also discuss some novel insights into the molecular basis and chemical perspectives of controlling Pol II transcriptional fidelity through structural, computational, and chemical biology approaches.
Keywords: Chemical biology; Computational biology; DNA damage recognition; Fidelity control; Nonpolar isostere; RNA polymerase II; Structural biology; Synthetic nucleotide analogs; Transcription elongation; Transcriptional lesion bypass; Unlocked nucleic acid.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Molecular basis of transcriptional pausing, stalling, and transcription-coupled repair initiation.Biochim Biophys Acta Gene Regul Mech. 2021 Jan;1864(1):194659. doi: 10.1016/j.bbagrm.2020.194659. Epub 2020 Nov 30. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 33271312 Free PMC article. Review.
-
RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications.Crit Rev Biochem Mol Biol. 2015;50(6):503-19. doi: 10.3109/10409238.2015.1087960. Epub 2015 Sep 22. Crit Rev Biochem Mol Biol. 2015. PMID: 26392149 Free PMC article. Review.
-
Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity.J Am Chem Soc. 2012 May 16;134(19):8231-40. doi: 10.1021/ja302077d. Epub 2012 May 2. J Am Chem Soc. 2012. PMID: 22509745 Free PMC article.
-
Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis.Proc Natl Acad Sci U S A. 2017 Aug 22;114(34):E7082-E7091. doi: 10.1073/pnas.1708748114. Epub 2017 Aug 7. Proc Natl Acad Sci U S A. 2017. PMID: 28784758 Free PMC article.
-
Strand-specific (asymmetric) contribution of phosphodiester linkages on RNA polymerase II transcriptional efficiency and fidelity.Proc Natl Acad Sci U S A. 2014 Aug 12;111(32):E3269-76. doi: 10.1073/pnas.1406234111. Epub 2014 Jul 29. Proc Natl Acad Sci U S A. 2014. PMID: 25074911 Free PMC article.
Cited by
-
Peripheral Immune Cells Exhaustion and Functional Impairment in Patients With Chronic Hepatitis B.Front Med (Lausanne). 2021 Oct 26;8:759292. doi: 10.3389/fmed.2021.759292. eCollection 2021. Front Med (Lausanne). 2021. PMID: 34782855 Free PMC article.
-
Functional interplay between NTP leaving group and base pair recognition during RNA polymerase II nucleotide incorporation revealed by methylene substitution.Nucleic Acids Res. 2016 May 5;44(8):3820-8. doi: 10.1093/nar/gkw220. Epub 2016 Apr 7. Nucleic Acids Res. 2016. PMID: 27060150 Free PMC article.
-
RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.Proc Natl Acad Sci U S A. 2022 Jan 18;119(3):e2114065119. doi: 10.1073/pnas.2114065119. Proc Natl Acad Sci U S A. 2022. PMID: 35022237 Free PMC article.
-
Molecular basis of transcriptional pausing, stalling, and transcription-coupled repair initiation.Biochim Biophys Acta Gene Regul Mech. 2021 Jan;1864(1):194659. doi: 10.1016/j.bbagrm.2020.194659. Epub 2020 Nov 30. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 33271312 Free PMC article. Review.
-
RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications.Crit Rev Biochem Mol Biol. 2015;50(6):503-19. doi: 10.3109/10409238.2015.1087960. Epub 2015 Sep 22. Crit Rev Biochem Mol Biol. 2015. PMID: 26392149 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

