An improved genome release (version Mt4.0) for the model legume Medicago truncatula
- PMID: 24767513
- PMCID: PMC4234490
- DOI: 10.1186/1471-2164-15-312
An improved genome release (version Mt4.0) for the model legume Medicago truncatula
Abstract
Background: Medicago truncatula, a close relative of alfalfa, is a preeminent model for studying nitrogen fixation, symbiosis, and legume genomics. The Medicago sequencing project began in 2003 with the goal to decipher sequences originated from the euchromatic portion of the genome. The initial sequencing approach was based on a BAC tiling path, culminating in a BAC-based assembly (Mt3.5) as well as an in-depth analysis of the genome published in 2011.
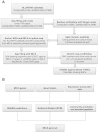
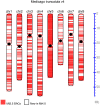
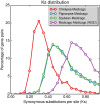
Results: Here we describe a further improved and refined version of the M. truncatula genome (Mt4.0) based on de novo whole genome shotgun assembly of a majority of Illumina and 454 reads using ALLPATHS-LG. The ALLPATHS-LG scaffolds were anchored onto the pseudomolecules on the basis of alignments to both the optical map and the genotyping-by-sequencing (GBS) map. The Mt4.0 pseudomolecules encompass ~360 Mb of actual sequences spanning 390 Mb of which ~330 Mb align perfectly with the optical map, presenting a drastic improvement over the BAC-based Mt3.5 which only contained 70% sequences (~250 Mb) of the current version. Most of the sequences and genes that previously resided on the unanchored portion of Mt3.5 have now been incorporated into the Mt4.0 pseudomolecules, with the exception of ~28 Mb of unplaced sequences. With regard to gene annotation, the genome has been re-annotated through our gene prediction pipeline, which integrates EST, RNA-seq, protein and gene prediction evidences. A total of 50,894 genes (31,661 high confidence and 19,233 low confidence) are included in Mt4.0 which overlapped with ~82% of the gene loci annotated in Mt3.5. Of the remaining genes, 14% of the Mt3.5 genes have been deprecated to an "unsupported" status and 4% are absent from the Mt4.0 predictions.
Conclusions: Mt4.0 and its associated resources, such as genome browsers, BLAST-able datasets and gene information pages, can be found on the JCVI Medicago web site (http://www.jcvi.org/medicago). The assembly and annotation has been deposited in GenBank (BioProject: PRJNA10791). The heavily curated chromosomal sequences and associated gene models of Medicago will serve as a better reference for legume biology and comparative genomics.
Figures




References
-
- Young ND, Debelle F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, Van de Peer Y, Proost S, Cook DR, Meyers BC, Spannagl M, Cheung F, De Mita S, Krishnakumar V, Gundlach H, Zhou S, Mudge J, Bharti AK, Murray JD, Naoumkina MA, Rosen B, Silverstein KA, Tang H, Rombauts S, Zhao PX, Zhou P. et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. - PMC - PubMed
-
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6(1):4. doi: 10.1186/1939-8433-6-4. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

