Analysis of surface-exposed outer membrane proteins in Helicobacter pylori
- PMID: 24769695
- PMCID: PMC4054173
- DOI: 10.1128/JB.01768-14
Analysis of surface-exposed outer membrane proteins in Helicobacter pylori
Abstract
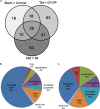
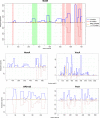
More than 50 Helicobacter pylori genes are predicted to encode outer membrane proteins (OMPs), but there has been relatively little experimental investigation of the H. pylori cell surface proteome. In this study, we used selective biotinylation to label proteins localized to the surface of H. pylori, along with differential detergent extraction procedures to isolate proteins localized to the outer membrane. Proteins that met multiple criteria for surface-exposed outer membrane localization included known adhesins, as well as Cag proteins required for activity of the cag type IV secretion system, putative lipoproteins, and other proteins not previously recognized as cell surface components. We identified sites of nontryptic cleavage consistent with signal sequence cleavage, as well as C-terminal motifs that may be important for protein localization. A subset of surface-exposed proteins were highly susceptible to proteolysis when intact bacteria were treated with proteinase K. Most Hop and Hom OMPs were susceptible to proteolysis, whereas Hor and Hof proteins were relatively resistant. Most of the protease-susceptible OMPs contain a large protease-susceptible extracellular domain exported beyond the outer membrane and a protease-resistant domain at the C terminus with a predicted β-barrel structure. These features suggest that, similar to the secretion of the VacA passenger domain, the N-terminal domains of protease-susceptible OMPs are exported through an autotransporter pathway. Collectively, these results provide new insights into the repertoire of surface-exposed H. pylori proteins that may mediate bacterium-host interactions, as well as the cell surface topology of these proteins.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

