Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification
- PMID: 24770324
- PMCID: PMC4263420
- DOI: 10.1038/nbt.2909
Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification
Abstract
Genome editing by Cas9, which cleaves double-stranded DNA at a sequence programmed by a short single-guide RNA (sgRNA), can result in off-target DNA modification that may be detrimental in some applications. To improve DNA cleavage specificity, we generated fusions of catalytically inactive Cas9 and FokI nuclease (fCas9). DNA cleavage by fCas9 requires association of two fCas9 monomers that simultaneously bind target sites ∼15 or 25 base pairs apart. In human cells, fCas9 modified target DNA sites with >140-fold higher specificity than wild-type Cas9 and with an efficiency similar to that of paired Cas9 'nickases', recently engineered variants that cleave only one DNA strand per monomer. The specificity of fCas9 was at least fourfold higher than that of paired nickases at loci with highly similar off-target sites. Target sites that conform to the substrate requirements of fCas9 occur on average every 34 bp in the human genome, suggesting the versatility of this approach for highly specific genome-wide editing.
Figures
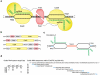

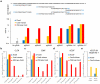

Comment in
-
More specific CRISPR editing.Nat Methods. 2014 Jul;11(7):712. doi: 10.1038/nmeth.3020. Nat Methods. 2014. PMID: 25110782 No abstract available.
Similar articles
-
Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing.Nat Biotechnol. 2014 Jun;32(6):569-76. doi: 10.1038/nbt.2908. Epub 2014 Apr 25. Nat Biotechnol. 2014. PMID: 24770325 Free PMC article.
-
Re-engineered RNA-Guided FokI-Nucleases for Improved Genome Editing in Human Cells.Mol Ther. 2017 Feb 1;25(2):342-355. doi: 10.1016/j.ymthe.2016.11.007. Mol Ther. 2017. PMID: 28153087 Free PMC article.
-
Generation of mutant mice via the CRISPR/Cas9 system using FokI-dCas9.Sci Rep. 2015 Jun 9;5:11221. doi: 10.1038/srep11221. Sci Rep. 2015. PMID: 26057433 Free PMC article.
-
[CRISPR/CAS9, the King of Genome Editing Tools].Mol Biol (Mosk). 2017 Jul-Aug;51(4):582-594. doi: 10.7868/S0026898417040036. Mol Biol (Mosk). 2017. PMID: 28900076 Review. Russian.
-
CRISPR/Cas9 in Genome Editing and Beyond.Annu Rev Biochem. 2016 Jun 2;85:227-64. doi: 10.1146/annurev-biochem-060815-014607. Epub 2016 Apr 25. Annu Rev Biochem. 2016. PMID: 27145843 Review.
Cited by
-
Undetectable off-target effects induced by FokI catalytic domain in mouse embryos.Genome Biol. 2024 Feb 20;25(1):51. doi: 10.1186/s13059-024-03188-9. Genome Biol. 2024. PMID: 38378658 Free PMC article.
-
Specificity of oligonucleotide gene therapy (OGT) agents.Theranostics. 2022 Oct 9;12(16):7132-7157. doi: 10.7150/thno.77830. eCollection 2022. Theranostics. 2022. PMID: 36276652 Free PMC article. Review.
-
Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing.Cells. 2020 Jul 2;9(7):1608. doi: 10.3390/cells9071608. Cells. 2020. PMID: 32630835 Free PMC article. Review.
-
Trans-spliced Cas9 allows cleavage of HBB and CCR5 genes in human cells using compact expression cassettes.Sci Rep. 2015 Jul 1;5:10777. doi: 10.1038/srep10777. Sci Rep. 2015. PMID: 26126518 Free PMC article.
-
Combining Off-flow, a Nextflow-coded program, and whole genome sequencing reveals unintended genetic variation in CRISPR/Cas-edited iPSCs.Comput Struct Biotechnol J. 2023 Dec 29;23:638-647. doi: 10.1016/j.csbj.2023.12.036. eCollection 2024 Dec. Comput Struct Biotechnol J. 2023. PMID: 38283851 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

