Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) dunal
- PMID: 24770414
- PMCID: PMC4059165
- DOI: 10.1074/jbc.M114.571919
Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) dunal
Abstract
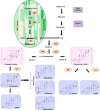
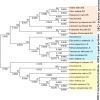
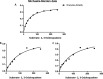
Oxidosqualene cyclases (OSCs) positioned at a key metabolic subdividing junction execute indispensable enzymatic cyclization of 2,3-oxidosqualene for varied triterpenoid biosynthesis. Such branch points present favorable gene targets for redirecting metabolic flux toward specific secondary metabolites. However, detailed information regarding the candidate OSCs covering different branches and their regulation is necessary for the desired genetic manipulation. The aim of the present study, therefore, was to characterize members of OSC superfamily from Withania somnifera (Ws), a medicinal plant of immense repute known to synthesize a large array of biologically active steroidal lactone triterpenoids called withanolides. Three full-length OSC cDNAs, β-amyrin synthase (WsOSC/BS), lupeol synthase (WsOSC/LS), and cycloartenol synthase (WsOSC/CS), having open reading frames of 2289, 2268, and 2277 bp, were isolated. Heterologous expression in Schizosaccharomyces pombe, LC-MS analyses, and kinetic studies confirmed their monofunctionality. The three WsOSCs were found to be spatially regulated at transcriptional level with WsOSC/CS being maximally expressed in leaf tissue. Promoter analysis of three WsOSCs genes resulted in identification of distinct cis-regulatory elements. Further, transcript profiling under methyl jasmonate, gibberellic acid, and yeast extract elicitations displayed differential transcriptional regulation of each of the OSCs. Changes were also observed in mRNA levels under elicitations and further substantiated with protein expression levels by Western blotting. Negative regulation by yeast extract resulted in significant increase in withanolide content. Empirical evidence suggests that repression of competitive branch OSCs like WsOSC/BS and WsOSC/LS possibly leads to diversion of substrate pool toward WsOSC/CS for increased withanolide production.
Keywords: Gene Regulation; Isoprenoid; Mass Spectrometry (MS); Metabolic Engineering; Western Blot.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures









References
-
- Phillips D. R., Rasbery J. M., Bartel B., Matsuda S. P. (2006) Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314 - PubMed
-
- Mahato S. B., Nandy A. K., Roy G. (1992) Triterpenoids. Phytochemistry 31, 2199–2249 - PubMed
-
- Lattoo S. K., Dhar R. S., Khan S., Bamotra S., Dhar A. K. (2007) Temporal sexual maturation and incremental stamina movement encourages mixed mating in Withania somnifera: an insurance for reproductive success. Curr. Sci. 92, 1390–1399
-
- Elsakka M., Grigorescu E., Stanescu U., Dorneanu V. (1990) New data referring to chemistry of Withania somnifera species. Rev. Med. Chir. Soc. Med. Nat. Iasi. 94, 385–387 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

