Protein phosphatase 2A (PP2A) regulates low density lipoprotein uptake through regulating sterol response element-binding protein-2 (SREBP-2) DNA binding
- PMID: 24770487
- PMCID: PMC4059166
- DOI: 10.1074/jbc.M114.570390
Protein phosphatase 2A (PP2A) regulates low density lipoprotein uptake through regulating sterol response element-binding protein-2 (SREBP-2) DNA binding
Abstract
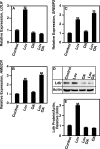
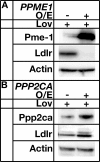
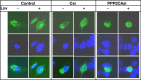
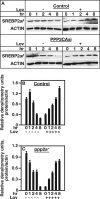
LDL-cholesterol (LDL-C) uptake by Ldlr is regulated at the transcriptional level by the cleavage-dependent activation of membrane-associated sterol response element-binding protein (SREBP-2). Activated SREBP-2 translocates to the nucleus, where it binds to an LDLR promoter sterol response element (SRE), increasing LDLR gene expression and LDL-C uptake. SREBP-2 cleavage and translocation steps are well established. Several SREBP-2 phosphorylation sites have been mapped and functionally characterized. The phosphatases dephosphorylating these sites remain elusive. The phosphatase(s) regulating SREBP-2 represents a novel pharmacological target for treating hypercholesterolemia. Here we show that protein phosphatase 2A (PP2A) promotes SREBP-2 LDLR promoter binding in response to cholesterol depletion. No binding to an LDLR SRE was observed in the presence of the HMG-CoA reductase inhibitor, lovastatin, when PP2A activity was inhibited by okadaic acid or depleted by siRNA methods. SREBP-2 cleavage and nuclear translocation were not affected by loss of PP2A. PP2A activity was required for SREBP-2 DNA binding. In response to cholesterol depletion, PP2A directly interacted with SREBP-2 and altered its phosphorylation state, causing an increase in SREBP-2 binding to an LDLR SRE site. Increased binding resulted in induced LDLR gene expression and increased LDL uptake. We conclude that PP2A activity regulates cholesterol homeostasis and LDL-C uptake.
Keywords: Cholesterol; DNA; Lipid; Lipoprotein; Low Density Lipoprotein (LDL); Metabolism; SREBP; Transcription.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







Similar articles
-
Alginate oligosaccharide enhances LDL uptake via regulation of LDLR and PCSK9 expression.J Nutr Biochem. 2015 Nov;26(11):1393-400. doi: 10.1016/j.jnutbio.2015.07.009. Epub 2015 Jul 30. J Nutr Biochem. 2015. PMID: 26320675
-
MK-2206, an allosteric inhibitor of AKT, stimulates LDLR expression and LDL uptake: A potential hypocholesterolemic agent.Atherosclerosis. 2018 Sep;276:28-38. doi: 10.1016/j.atherosclerosis.2018.07.009. Epub 2018 Jul 7. Atherosclerosis. 2018. PMID: 30025252
-
Resveratrol increases the expression and activity of the low density lipoprotein receptor in hepatocytes by the proteolytic activation of the sterol regulatory element-binding proteins.Atherosclerosis. 2012 Feb;220(2):369-74. doi: 10.1016/j.atherosclerosis.2011.11.006. Epub 2011 Nov 16. Atherosclerosis. 2012. PMID: 22153697
-
Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins.World J Gastroenterol. 2004 Nov 1;10(21):3081-7. doi: 10.3748/wjg.v10.i21.3081. World J Gastroenterol. 2004. PMID: 15457548 Free PMC article. Review.
-
Targeting the proprotein convertase subtilisin/kexin type 9 for the treatment of dyslipidemia and atherosclerosis.J Am Coll Cardiol. 2013 Oct 15;62(16):1401-8. doi: 10.1016/j.jacc.2013.07.056. Epub 2013 Aug 21. J Am Coll Cardiol. 2013. PMID: 23973703 Review.
Cited by
-
Inhibition of AMP Kinase by the Protein Phosphatase 2A Heterotrimer, PP2APpp2r2d.J Biol Chem. 2015 Apr 24;290(17):10588-98. doi: 10.1074/jbc.M114.626259. Epub 2015 Feb 18. J Biol Chem. 2015. PMID: 25694423 Free PMC article.
-
Prebiotics Beyond the Gut: Omics Insights, Artificial Intelligence, and Clinical Trials in Organ-Specific Applications.Probiotics Antimicrob Proteins. 2025 Aug;17(4):2500-2521. doi: 10.1007/s12602-025-10465-x. Epub 2025 Jan 29. Probiotics Antimicrob Proteins. 2025. PMID: 39878922 Review.
-
Quercetin improves lipid metabolism via SCAP-SREBP2-LDLr signaling pathway in early stage diabetic nephropathy.Diabetes Metab Syndr Obes. 2019 May 30;12:827-839. doi: 10.2147/DMSO.S195456. eCollection 2019. Diabetes Metab Syndr Obes. 2019. PMID: 31239739 Free PMC article.
-
Emerging risk biomarkers in cardiovascular diseases and disorders.J Lipids. 2015;2015:971453. doi: 10.1155/2015/971453. Epub 2015 Apr 8. J Lipids. 2015. PMID: 25949827 Free PMC article. Review.
-
Simvastatin reduces TGF-β1-induced SMAD2/3-dependent human ventricular fibroblasts differentiation: Role of protein phosphatase activation.Int J Cardiol. 2018 Nov 1;270:228-236. doi: 10.1016/j.ijcard.2018.06.061. Epub 2018 Jun 21. Int J Cardiol. 2018. PMID: 30220377 Free PMC article.
References
-
- Maxfield F. R., Tabas I. (2005) Role of cholesterol and lipid and organization in disease. Nature 438, 612–621 - PubMed
-
- Sonnino S., Prinetti A. (2013) Membrane domains and the “lipid raft” concept. Curr. Med. Chem. 20, 4–21 - PubMed
-
- Incardona J. P., Eaton S. (2000) Cholesterol in Signal Transduction. Curr. Opin. Cell Biol. 12, 193–203 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

