A protease storm cleaves a cell-cell adhesion molecule in cancer: multiple proteases converge to regulate PTPmu in glioma cells
- PMID: 24771611
- PMCID: PMC4600327
- DOI: 10.1002/jcb.24824
A protease storm cleaves a cell-cell adhesion molecule in cancer: multiple proteases converge to regulate PTPmu in glioma cells
Abstract
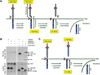
Cleavage of the cell-cell adhesion molecule, PTPµ, occurs in human glioblastoma multiforme brain tumor tissue and glioma cell lines. PTPµ cleavage is linked to increased cell motility and growth factor independent survival of glioma cells in vitro. Previously, PTPµ was shown to be cleaved by furin in the endoplasmic reticulum to generate membrane associated E- (extracellular) and P- (phosphatase) subunits, and by ADAMs and the gamma secretase complex at the plasma membrane. We also identified the presence of additional extracellular and intracellular PTPµ fragments in brain tumors. We set out to biochemically analyze PTPµ cleavage in cancer cells. We determined that, in addition to the furin-processed form of PTPµ, a pool of 200 kDa full-length PTPµ exists at the plasma membrane that is cleaved directly by ADAM to generate a larger shed form of the PTPµ extracellular segment. Notably, in glioma cells, full-length PTPµ is also subject to calpain cleavage, which generates novel PTPµ fragments not found in other immortalized cells. We also observed glycosylation and phosphorylation differences in the cancer cells. Our data suggest that an additional serine protease also contributes to PTPµ shedding in glioma cells. We hypothesize that a "protease storm" occurs in cancer cells whereby multiple proteases converge to reduce the presence of cell-cell adhesion molecules at the plasma membrane and to generate protein fragments with unique biological functions. As a consequence, the "protease storm" could promote the migration and invasion of tumor cells.
Keywords: PROTEOLYSIS; SHEDDING; PTPmu; RECEPTOR PROTEIN TYROSINE PHOSPHATASE; PROTEIN TYROSINE PHOSPHATASE; ADAM; CALPAIN; FURIN; SERINE PROTEASE; GLIOMA.
© 2014 Wiley Periodicals, Inc.
Figures





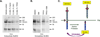
Similar articles
-
Identification of phospholipase C gamma1 as a protein tyrosine phosphatase mu substrate that regulates cell migration.J Cell Biochem. 2011 Jan;112(1):39-48. doi: 10.1002/jcb.22710. J Cell Biochem. 2011. PMID: 20506511 Free PMC article.
-
Protein tyrosine phosphatase mu regulates glioblastoma cell growth and survival in vivo.Neuro Oncol. 2012 May;14(5):561-73. doi: 10.1093/neuonc/nos066. Epub 2012 Apr 14. Neuro Oncol. 2012. PMID: 22505657 Free PMC article.
-
Artificial Intelligence-Based Computational Screening and Functional Assays Identify Candidate Small Molecule Antagonists of PTPmu-Dependent Adhesion.Int J Mol Sci. 2023 Feb 21;24(5):4274. doi: 10.3390/ijms24054274. Int J Mol Sci. 2023. PMID: 36901713 Free PMC article.
-
Signaling Determinants of Glioma Cell Invasion.Adv Exp Med Biol. 2020;1202:129-149. doi: 10.1007/978-3-030-30651-9_7. Adv Exp Med Biol. 2020. PMID: 32034712 Review.
-
Protein tyrosine phosphatase receptor type kappa (PTPRK) revisited: evolving insights into structure, function, and pathology.J Transl Med. 2025 May 12;23(1):534. doi: 10.1186/s12967-025-06496-1. J Transl Med. 2025. PMID: 40355891 Free PMC article. Review.
Cited by
-
PTPRM, a candidate tumor suppressor gene in small intestinal neuroendocrine tumors.Endocr Connect. 2019 Aug 1;8(8):1126-1135. doi: 10.1530/EC-19-0279. Endocr Connect. 2019. PMID: 31349215 Free PMC article.
-
Protein tyrosine phosphatases: promising targets in pancreatic ductal adenocarcinoma.Cell Mol Life Sci. 2019 Jul;76(13):2571-2592. doi: 10.1007/s00018-019-03095-4. Epub 2019 Apr 13. Cell Mol Life Sci. 2019. PMID: 30982078 Free PMC article. Review.
-
SLPI is a critical mediator that controls PTH-induced bone formation.Nat Commun. 2021 Apr 9;12(1):2136. doi: 10.1038/s41467-021-22402-x. Nat Commun. 2021. PMID: 33837198 Free PMC article.
-
NF-κB inhibitor with Temozolomide results in significant apoptosis in glioblastoma via the NF-κB(p65) and actin cytoskeleton regulatory pathways.Sci Rep. 2020 Aug 7;10(1):13352. doi: 10.1038/s41598-020-70392-5. Sci Rep. 2020. PMID: 32770097 Free PMC article.
-
PTPmu-targeted nanoparticles label invasive pediatric and adult glioblastoma.Nanomedicine. 2020 Aug;28:102216. doi: 10.1016/j.nano.2020.102216. Epub 2020 May 13. Nanomedicine. 2020. PMID: 32413511 Free PMC article.
References
-
- Aricescu AR, Fulga TA, Cismasiu V, Goody RS, Szedlacsek SE. Intramolecular interactions in protein tyrosine phosphatase RPTPmu: kinetic evidence. Biochem Biophys Res Commun. 2001;280:319–327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

