Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice
- PMID: 24771859
- PMCID: PMC4055923
- DOI: 10.1182/blood-2013-08-521252
Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice
Abstract
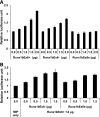
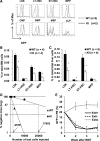
RUNX1 is an important transcription factor for hematopoiesis. There are multiple alternatively spliced isoforms of RUNX1. The best known isoforms are RUNX1a from use of exon 7A and RUNX1b and c from use of exon 7B. RUNX1a has unique functions due to its lack of C-terminal regions common to RUNX1b and c. Here, we report that the ortholog of human RUNX1a was only found in primates. Furthermore, we characterized 3 Runx1 isoforms generated by exon 6 alternative splicing. Runx1bEx6(-) (Runx1b without exon 6) and a unique mouse Runx1bEx6e showed higher colony-forming activity than the full-length Runx1b (Runx1bEx6(+)). They also facilitated the transactivation of Runx1bEx6(+). To gain insight into in vivo functions, we analyzed a knock-in (KI) mouse model that lacks isoforms Runx1b/cEx6(-) and Runx1bEx6e. KI mice had significantly fewer lineage-Sca1(+)c-Kit(+) cells, short-term hematopoietic stem cells (HSCs) and multipotent progenitors than controls. In vivo competitive repopulation assays demonstrated a sevenfold difference of functional HSCs between wild-type and KI mice. Together, our results show that Runx1 isoforms involving exon 6 support high self-renewal capacity in vitro, and their loss results in reduction of the HSC pool in vivo, which underscore the importance of fine-tuning RNA splicing in hematopoiesis.
© 2014 by The American Society of Hematology.
Figures




Comment in
-
An unsung runt 6e isoform for HSC expansion.Blood. 2014 Jun 12;123(24):3684-6. doi: 10.1182/blood-2014-05-572891. Blood. 2014. PMID: 24926066 No abstract available.
References
-
- Takahashi A, Satake M, Yamaguchi-Iwai Y, et al. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86(2):607–616. - PubMed
-
- Huang G, Zhang P, Hirai H, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40(1):51–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

