Structural and functional insights to ubiquitin-like protein conjugation
- PMID: 24773014
- PMCID: PMC4118471
- DOI: 10.1146/annurev-biophys-051013-022958
Structural and functional insights to ubiquitin-like protein conjugation
Abstract
Attachment of ubiquitin (Ub) and ubiquitin-like proteins (Ubls) to cellular proteins regulates numerous cellular processes including transcription, the cell cycle, stress responses, DNA repair, apoptosis, immune responses, and autophagy, to name a few. The mechanistically parallel but functionally distinct conjugation pathways typically require the concerted activities of three types of protein: E1 Ubl-activating enzymes, E2 Ubl carrier proteins, and E3 Ubl ligases. E1 enzymes initiate pathway specificity for each cascade by recognizing and activating cognate Ubls, followed by catalyzing Ubl transfer to cognate E2 protein(s). Under certain circumstances, the E2 Ubl complex can direct ligation to the target protein, but most often requires the cooperative activity of E3 ligases. Reviewed here are recent structural and functional studies that improve our mechanistic understanding of E1-, E2-, and E3-mediated Ubl conjugation.
Keywords: E1; E2; E3; ligase; ubiquitin; ubiquitin-like protein.
Figures
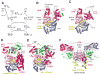
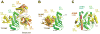
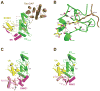
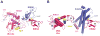
References
-
- Aravind L, Koonin EV. The U box is a modified RING finger---a common domain in ubiquitination. Curr Biol. 2000;10:R132–34. - PubMed
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. - PubMed
-
- Boh BK, Smith PG, Hagen T. Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J Mol Biol. 2011;409:136–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

