SMOQ: a tool for predicting the absolute residue-specific quality of a single protein model with support vector machines
- PMID: 24776231
- PMCID: PMC4013430
- DOI: 10.1186/1471-2105-15-120
SMOQ: a tool for predicting the absolute residue-specific quality of a single protein model with support vector machines
Abstract
Background: It is important to predict the quality of a protein structural model before its native structure is known. The method that can predict the absolute local quality of individual residues in a single protein model is rare, yet particularly needed for using, ranking and refining protein models.
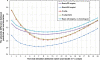
Results: We developed a machine learning tool (SMOQ) that can predict the distance deviation of each residue in a single protein model. SMOQ uses support vector machines (SVM) with protein sequence and structural features (i.e. basic feature set), including amino acid sequence, secondary structures, solvent accessibilities, and residue-residue contacts to make predictions. We also trained a SVM model with two new additional features (profiles and SOV scores) on 20 CASP8 targets and found that including them can only improve the performance when real deviations between native and model are higher than 5Å. The SMOQ tool finally released uses the basic feature set trained on 85 CASP8 targets. Moreover, SMOQ implemented a way to convert predicted local quality scores into a global quality score. SMOQ was tested on the 84 CASP9 single-domain targets. The average difference between the residue-specific distance deviation predicted by our method and the actual distance deviation on the test data is 2.637Å. The global quality prediction accuracy of the tool is comparable to other good tools on the same benchmark.
Conclusion: SMOQ is a useful tool for protein single model quality assessment. Its source code and executable are available at: http://sysbio.rnet.missouri.edu/multicom_toolbox/.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

