Signal amplification and transduction in phytochrome photosensors
- PMID: 24776794
- PMCID: PMC4015848
- DOI: 10.1038/nature13310
Signal amplification and transduction in phytochrome photosensors
Abstract
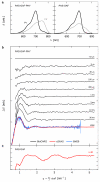
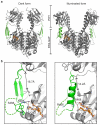
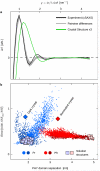
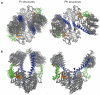
Sensory proteins must relay structural signals from the sensory site over large distances to regulatory output domains. Phytochromes are a major family of red-light-sensing kinases that control diverse cellular functions in plants, bacteria and fungi. Bacterial phytochromes consist of a photosensory core and a carboxy-terminal regulatory domain. Structures of photosensory cores are reported in the resting state and conformational responses to light activation have been proposed in the vicinity of the chromophore. However, the structure of the signalling state and the mechanism of downstream signal relay through the photosensory core remain elusive. Here we report crystal and solution structures of the resting and activated states of the photosensory core of the bacteriophytochrome from Deinococcus radiodurans. The structures show an open and closed form of the dimeric protein for the activated and resting states, respectively. This nanometre-scale rearrangement is controlled by refolding of an evolutionarily conserved 'tongue', which is in contact with the chromophore. The findings reveal an unusual mechanism in which atomic-scale conformational changes around the chromophore are first amplified into an ångstrom-scale distance change in the tongue, and further grow into a nanometre-scale conformational signal. The structural mechanism is a blueprint for understanding how phytochromes connect to the cellular signalling network.
Figures
Comment in
-
Structural biology: Action at a distance in a light receptor.Nature. 2014 May 8;509(7499):174-5. doi: 10.1038/nature13331. Epub 2014 Apr 30. Nature. 2014. PMID: 24776796 No abstract available.
References
-
- Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273:1409–1412. doi:DOI 10.1126/science.273.5280.1409. - PubMed
-
- Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi:DOI 10.1126/science.277.5331.1505. - PubMed
-
- Jiang ZY, et al. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science. 1999;285:406–409. doi:DOI 10.1126/science.285.5426.406. - PubMed
Extended Data References
-
- Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7 doi: http://www.nature.com/msb/journal/v7/n1/suppinfo/msb201175_S1.html. - PMC - PubMed
-
- Wagner JR, Zhang JR, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. Journal of Biological Chemistry. 2007;282:12298–12309. doi:DOI 10.1074/jbc.M611824200. - PubMed
-
- Mailliet J, et al. Spectroscopy and a High-Resolution Crystal Structure of Tyr263 Mutants of Cyanobacterial Phytochrome Cph1. J. Mol. Biol. 2011;413:115–127. doi:DOI 10.1016/j.jmb.2011.08.023. - PubMed
-
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. doi:Doi 10.1107/S0021889803012779.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

