Astrocyte-encoded positional cues maintain sensorimotor circuit integrity
- PMID: 24776795
- PMCID: PMC4057936
- DOI: 10.1038/nature13161
Astrocyte-encoded positional cues maintain sensorimotor circuit integrity
Abstract
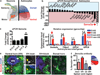
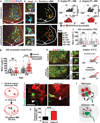
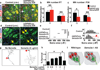
Astrocytes, the most abundant cells in the central nervous system, promote synapse formation and help to refine neural connectivity. Although they are allocated to spatially distinct regional domains during development, it is unknown whether region-restricted astrocytes are functionally heterogeneous. Here we show that postnatal spinal cord astrocytes express several region-specific genes, and that ventral astrocyte-encoded semaphorin 3a (Sema3a) is required for proper motor neuron and sensory neuron circuit organization. Loss of astrocyte-encoded Sema3a leads to dysregulated α-motor neuron axon initial segment orientation, markedly abnormal synaptic inputs, and selective death of α- but not of adjacent γ-motor neurons. In addition, a subset of TrkA(+) sensory afferents projects to ectopic ventral positions. These findings demonstrate that stable maintenance of a positional cue by developing astrocytes influences multiple aspects of sensorimotor circuit formation. More generally, they suggest that regional astrocyte heterogeneity may help to coordinate postnatal neural circuit refinement.
Figures











Comment in
-
Neural development: Astrocyte semaphores guide circuit formation.Nat Rev Neurosci. 2014 Jun;15(6):352-3. doi: 10.1038/nrn3757. Epub 2014 May 14. Nat Rev Neurosci. 2014. PMID: 24824178 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

